PROTEOMICS Tools Techniques Application KUMARI RASHMI SEM3 Introduction





- Slides: 18

PROTEOMICS Tools, Techniques & Application KUMARI RASHMI SEM-3

Introduction �Proteomics is the large-scale study of proteins, particularly their structures and function. �Proteomics is the systematic study of all of the proteins in a cell, tissue, or organism. �The term proteomics was coined in 1994 by Marc Wilkins. �The study of the Proteome. �The proteome is the entire set of proteome expressed by a genome, cell, tissue or organism.


Proteomics classification �Expression �Cell map or Interaction �Functional �Structural

Steps in Proteomic Research �Sample collection, handling and storage. �Protein seperation �Protein identification and characterization

Techniques used in Proteomics study � 2 -D gel electophoresis �ICAT(isotope coded affinity tags) �Mass spectroscopy �MALDI �PMF(Peptide mass fingerprinting)

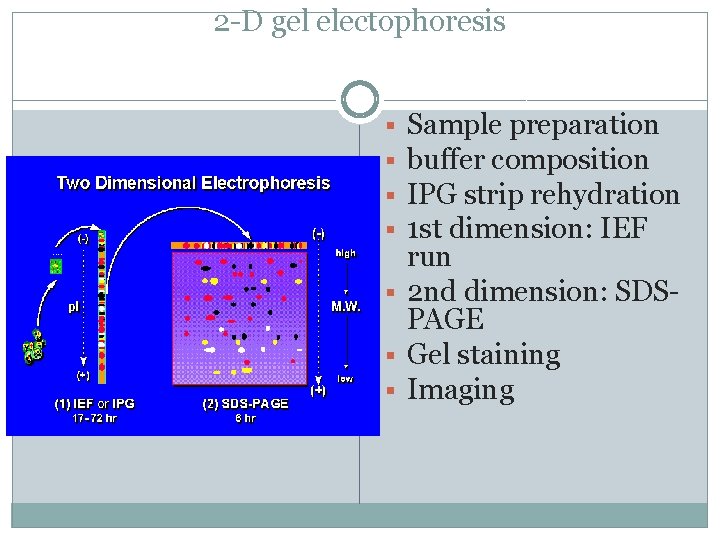
2 -D gel electophoresis Sample preparation buffer composition IPG strip rehydration 1 st dimension: IEF run § 2 nd dimension: SDSPAGE § Gel staining § Imaging § §

ICAT(isotope coded affinity tags) �Using ICAT you can compare relative protein abundance between two samples. �ICAT is particularly useful for comparision between healthy and diseased tissue

Mass spectroscopy �Used for protein identification. �Mass spectrometer seperates proteins according to their mass-to-charge (m/z) ratio. �used for determining masses of particles, for determining the elemental composition of a sample or molecule. �The molecule is first ionized which results in the formation of charged particles. The ions are separated according to their (m/z)ratio in an analyzer by electromagnetic fields.

MALDI Matrix-assisted laser desorption/ionization � is a soft ionization technique used in mass spectrometry, allowing the analysis of biomolecules and large organic molecules.

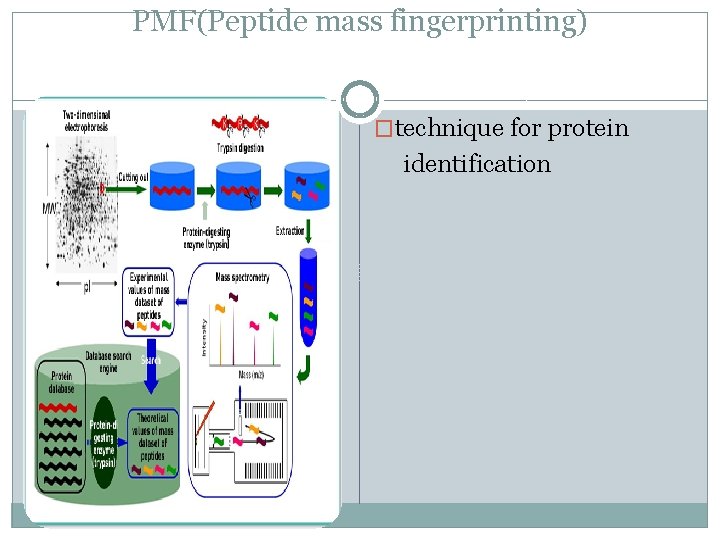
PMF(Peptide mass fingerprinting) �technique for protein identification

Database of proteomics �PRIDE-The PRIDE PRoteomics IDEntifications database is a centralised, standards compliant, public data repository for proteomics data. �PROTICdb access- PROTICdb is a tool developed by INRA that we have chosen for storing and processing our proteomic data. � The PPDB stores experimental data from in-house proteome and mass spectrometry analysis, curated information about protein function, protein properties and subcellular localization.

Human Proteome Project �Launched by the Human Proteome Organization (HUPO). � Designed to map the entire human protein set. �the three working pillars for HPP: Ø mass spectrometry, Ø antibodycapture, and Ø bioinformatics tools & knowledge bases.

Application of Proteomics � Nutrition Research �diabetes research. � Cardiovascular research. � Autoantibody profiling for the study and treatment of autoimmune disease. � Urological Cancer Research �Fetal and maternal medicine � Neurology � Renal disease diagnosis. � Study of Tumor Metastasis. �Protein Biomarker

Research Areas in Proteomics �Quantitative proteomics �Phosphoproteomics �Glycoproteomics �Proteogenomics �Neuroproteomics �Plant proteomics �Toxicoproteomics �Ethnoproteomics

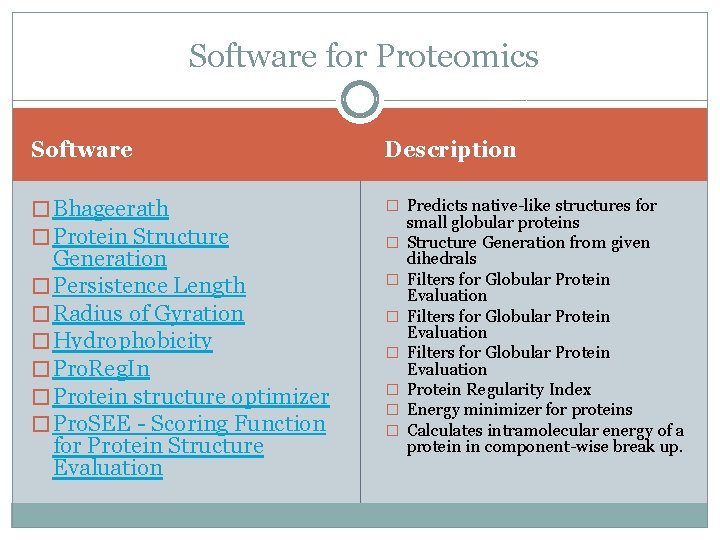
Software for Proteomics Software Description � Bhageerath � Protein Structure � Predicts native-like structures for Generation � Persistence Length � Radius of Gyration � Hydrophobicity � Pro. Reg. In � Protein structure optimizer � Pro. SEE - Scoring Function for Protein Structure Evaluation � � � � small globular proteins Structure Generation from given dihedrals Filters for Globular Protein Evaluation Protein Regularity Index Energy minimizer for proteins Calculates intramolecular energy of a protein in component-wise break up.


Thank You