Prokaryote Taxonomy Diversity Classification Nomenclature Identification Phenetic Numerical

- Slides: 22

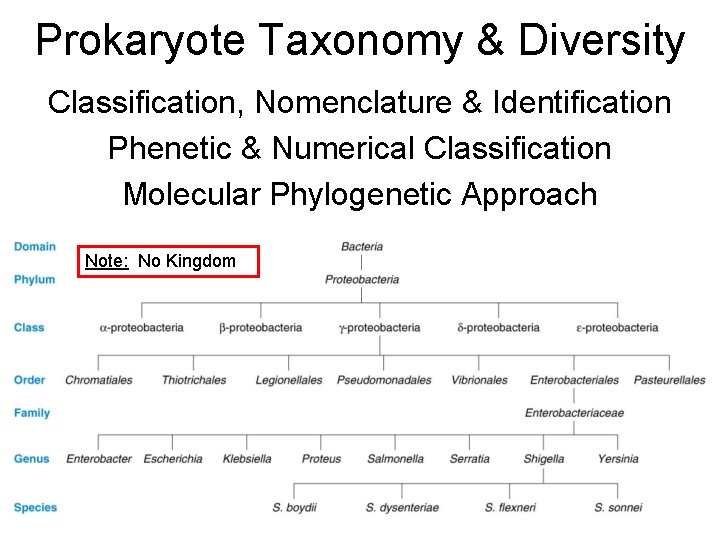
Prokaryote Taxonomy & Diversity Classification, Nomenclature & Identification Phenetic & Numerical Classification Molecular Phylogenetic Approach Note: No Kingdom

Basics to Know • A grouping based on similar characters is called a taxon. • Hierarchy of Prokaryote Taxa often omits Kingdom. Phylum often referred to as a Section. • Bergey’s Manual of Systematic Bacteriology – 1 st ed. (1989); mostly phenetic classification; 4 volumes – 2 nd ed. (2001 -2003); mostly phylogenetic classification; 5 volumes • Species defined differently for prokaryotes (no sexual reproduction) = a collection of genetically unique populations with many stable characters distinctly different from other groups. • Within a species there may be strains, populations characterized by minor variations in biochemical/ physiological properties (biovars), antigenic distinctions (serovars), shape (morphovars), or viral susceptibility (phage-typing).

The focus here is on best techniques for distinguishing strains of the same species.

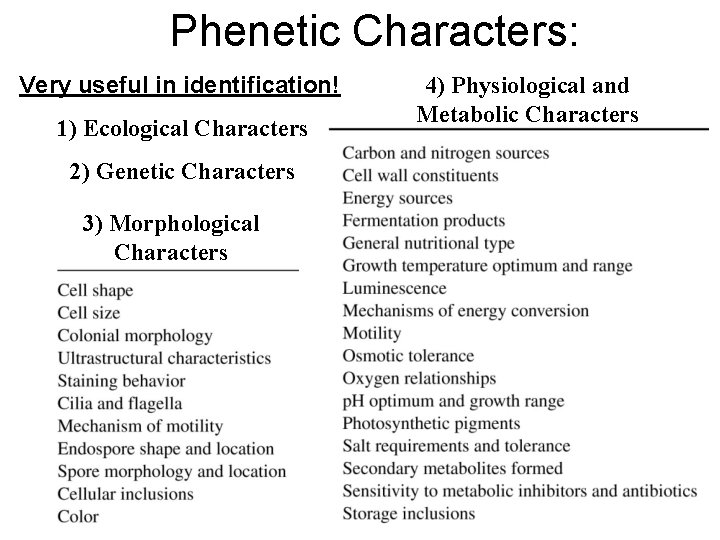
Phenetic Characters: Very useful in identification! 1) Ecological Characters 2) Genetic Characters 3) Morphological Characters 4) Physiological and Metabolic Characters

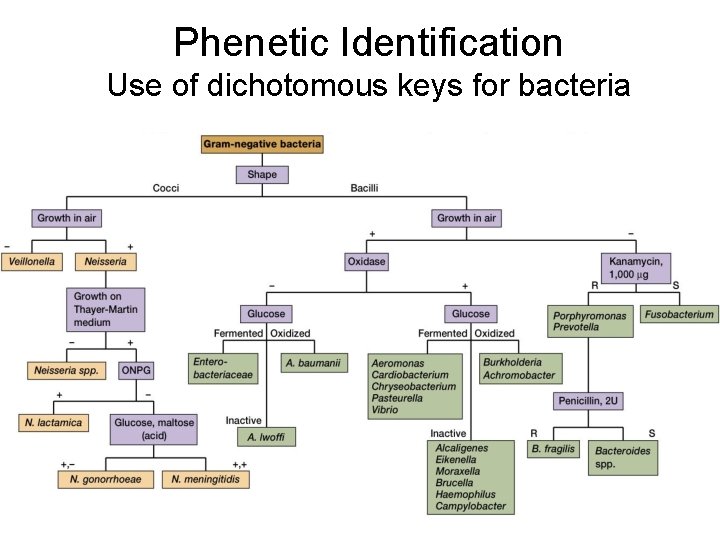
Phenetic Identification Use of dichotomous keys for bacteria

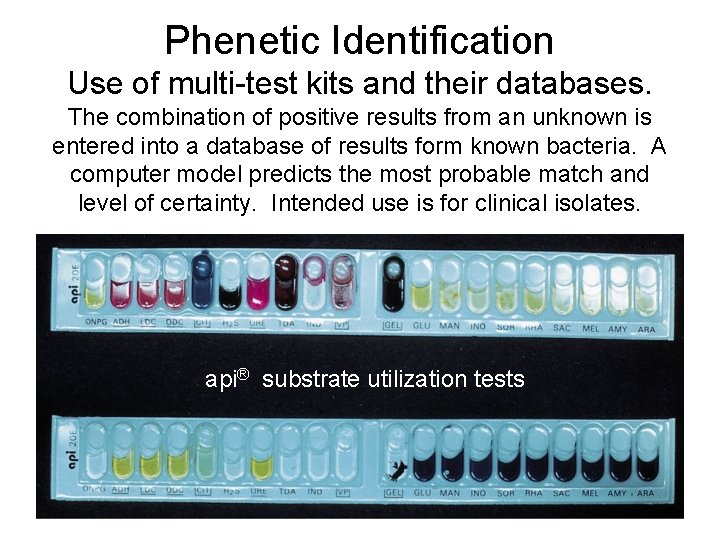
Phenetic Identification Use of multi-test kits and their databases. The combination of positive results from an unknown is entered into a database of results form known bacteria. A computer model predicts the most probable match and level of certainty. Intended use is for clinical isolates. api® substrate utilization tests

Molecular Characters • Fatty acid profiles (FAME analysis) • Proteins – Electrophoretic Mobility – Immuno-Reactivity – A. A. Sequence Data • Nucleic Acids – Nucleotide composition (G+C content ≈ Tm) – Degree of Hybridization (>70% ≈ species) – Nucleotide Sequence Data

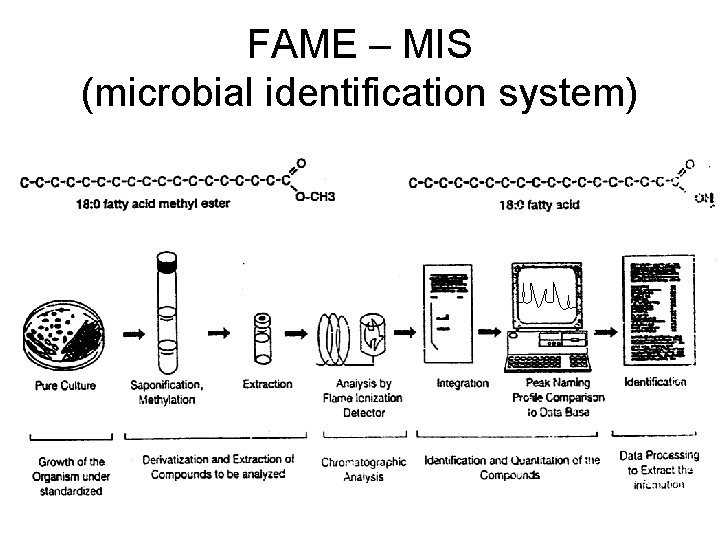
FAME – MIS (microbial identification system)

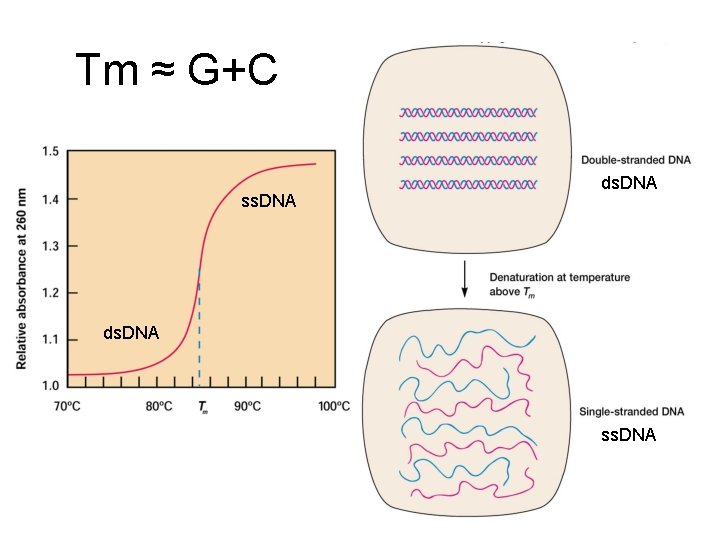
Tm ≈ G+C ss. DNA ds. DNA ss. DNA

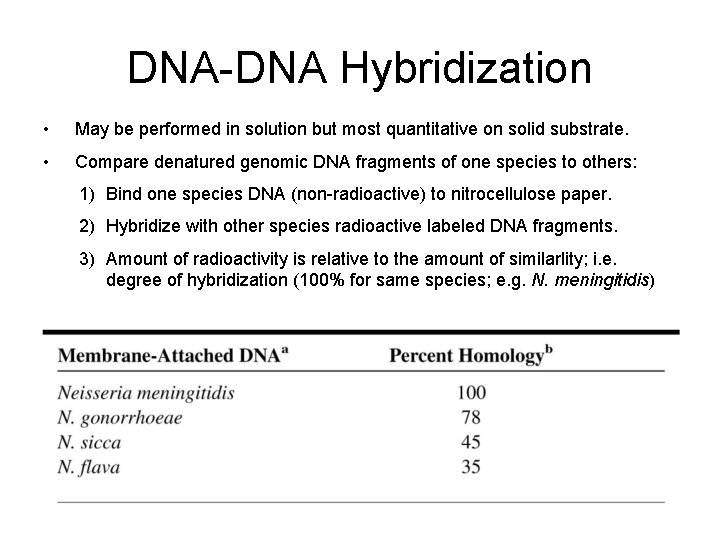
DNA-DNA Hybridization • May be performed in solution but most quantitative on solid substrate. • Compare denatured genomic DNA fragments of one species to others: 1) Bind one species DNA (non-radioactive) to nitrocellulose paper. 2) Hybridize with other species radioactive labeled DNA fragments. 3) Amount of radioactivity is relative to the amount of similarlity; i. e. degree of hybridization (100% for same species; e. g. N. meningitidis)

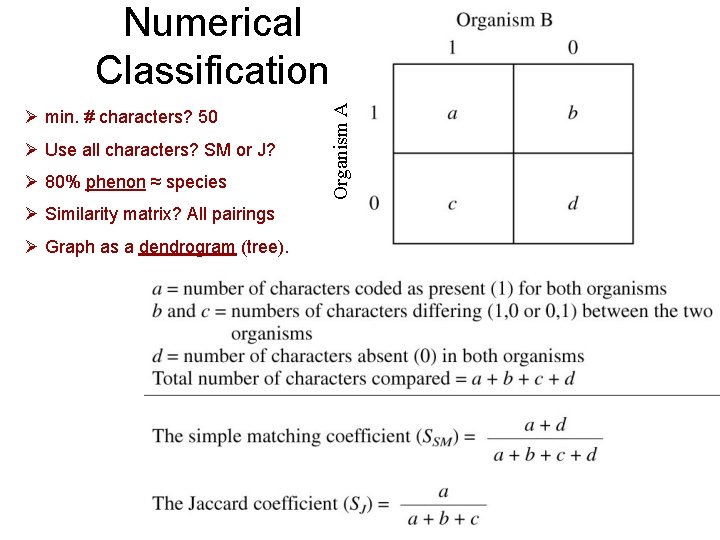
Ø min. # characters? 50 Ø Use all characters? SM or J? Ø 80% phenon ≈ species Ø Similarity matrix? All pairings Ø Graph as a dendrogram (tree). Organism A Numerical Classification

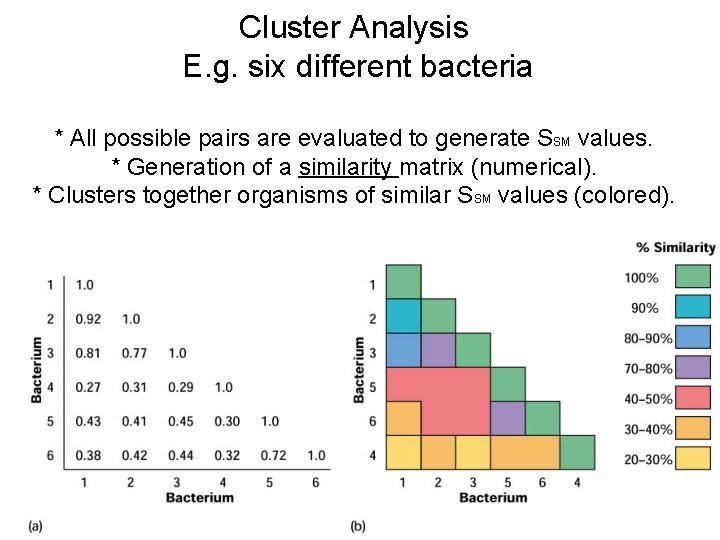
Cluster Analysis E. g. six different bacteria * All possible pairs are evaluated to generate SSM values. * Generation of a similarity matrix (numerical). * Clusters together organisms of similar SSM values (colored).

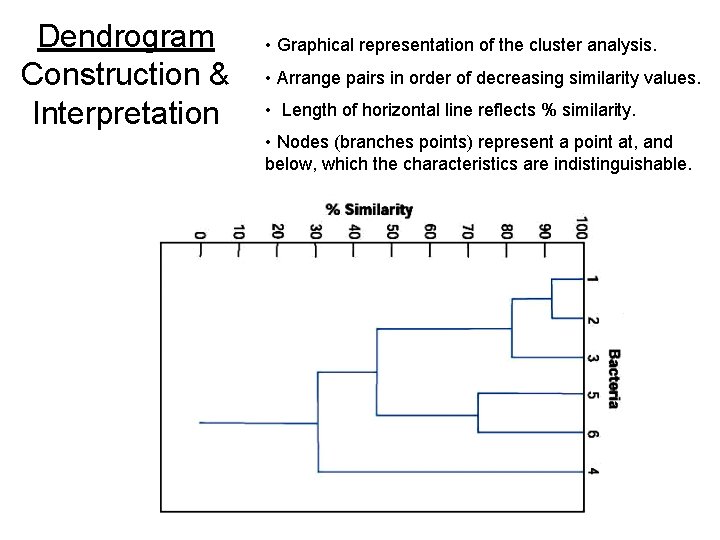
Dendrogram Construction & Interpretation • Graphical representation of the cluster analysis. • Arrange pairs in order of decreasing similarity values. • Length of horizontal line reflects % similarity. • Nodes (branches points) represent a point at, and below, which the characteristics are indistinguishable.

Phylogenetic Classification Molecular Chronometers • Phylogeny refers to grouping based on evolutionary relatedness; regardless of phenetic characters. • Phylogeny is inferred from changes in protein or DNA sequence over evolutionary time. • Attributes of an Ideal “Molecular Chronometer”: – – Universally distributed. Functionally homologous. Ease of analysis. Rate of sequence change commensurate with evolutionary distance measured.

Why the 16 Sr. RNA gene? • All cellular life has ribosomes with r. RNAs • All r. RNAs have the same function (transcription) in all cellular life. • The 16 Sr. RNA gene provides enough information (# sequence positions) and not too big that its cloning and sequencing is inefficient (time & $). • There are only 7 regions that are considered highly variable most of the molecule’s sequence is very conserved so to maintain critical functions.

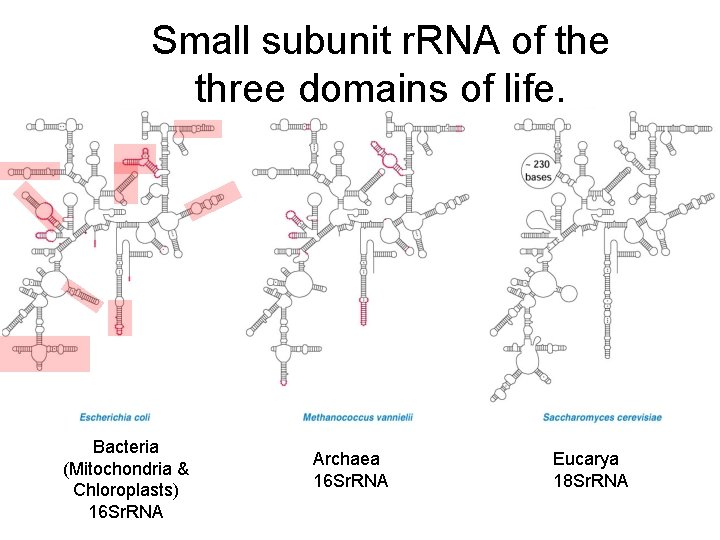
Small subunit r. RNA of the three domains of life. Bacteria (Mitochondria & Chloroplasts) 16 Sr. RNA Archaea 16 Sr. RNA Eucarya 18 Sr. RNA

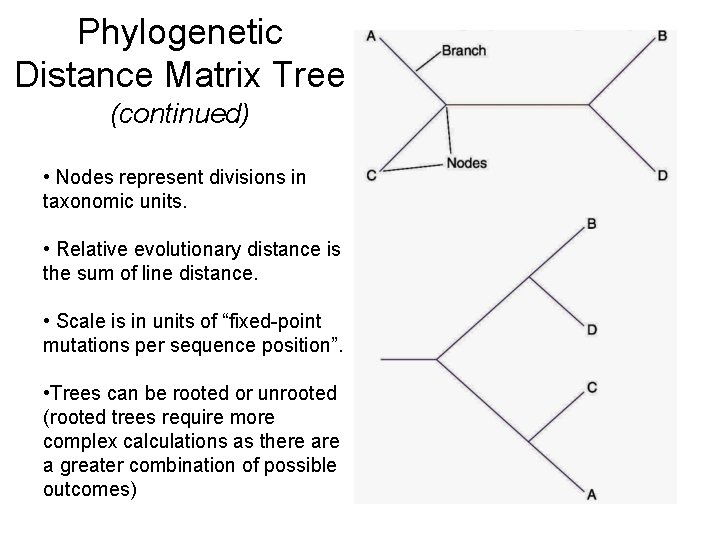
Phylogenetic Distance Matrix Tree • Sequence molecular chronometer of choice for all organisms studied. • Align sequences based on highly conserved sequence regions. • Pair-wise determination of differences. • Adjustments may be made for multiple mutations as particular sites. • Construction of tree requires massive computer calculations to determine the best fit of the data (similar to dendrogram).

Phylogenetic Distance Matrix Tree (continued) • Nodes represent divisions in taxonomic units. • Relative evolutionary distance is the sum of line distance. • Scale is in units of “fixed-point mutations per sequence position”. • Trees can be rooted or unrooted (rooted trees require more complex calculations as there a greater combination of possible outcomes)

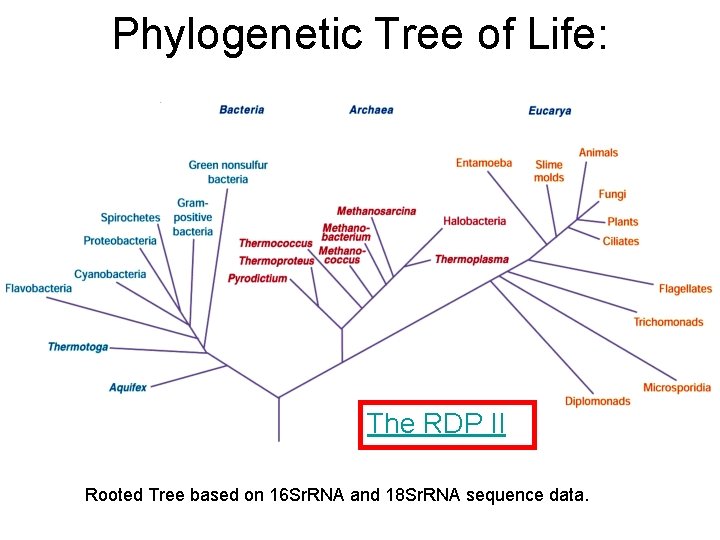
Phylogenetic Tree of Life: The RDP II Rooted Tree based on 16 Sr. RNA and 18 Sr. RNA sequence data.

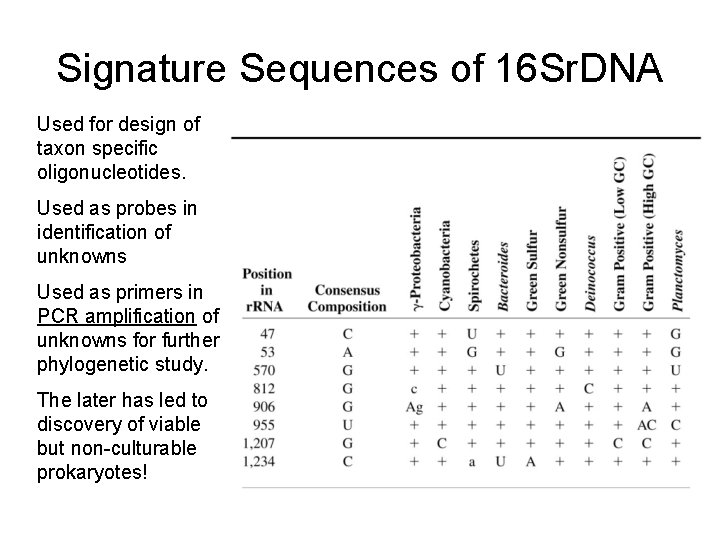
Signature Sequences of 16 Sr. DNA Used for design of taxon specific oligonucleotides. Used as probes in identification of unknowns Used as primers in PCR amplification of unknowns for further phylogenetic study. The later has led to discovery of viable but non-culturable prokaryotes!

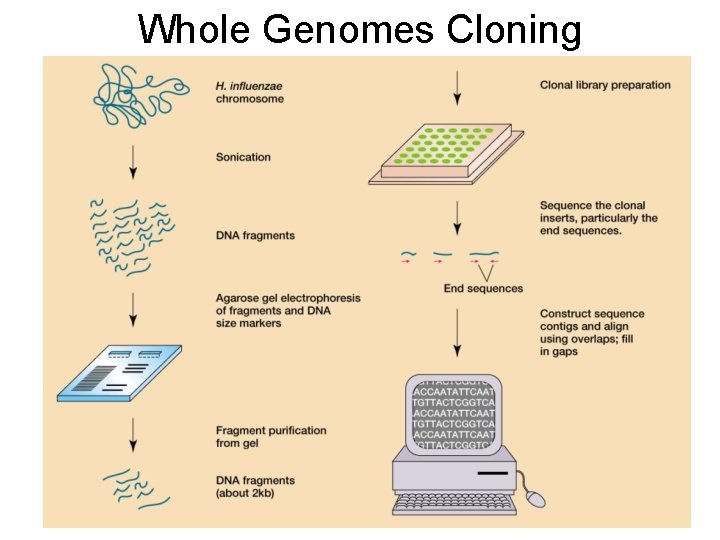
Whole Genomes Cloning

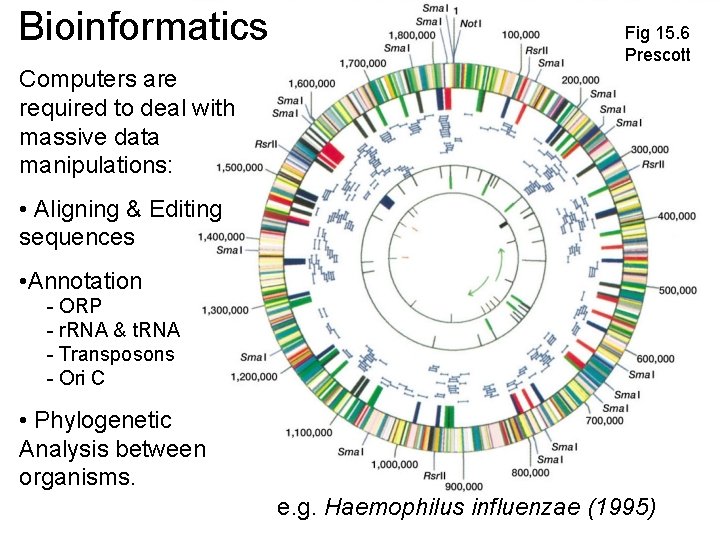
Bioinformatics Fig 15. 6 Prescott Computers are required to deal with massive data manipulations: • Aligning & Editing sequences • Annotation - ORP - r. RNA & t. RNA - Transposons - Ori C • Phylogenetic Analysis between organisms. e. g. Haemophilus influenzae (1995)