Prokaryote Taxonomy Diversity Classification Nomenclature Identification Phenetic Classification
- Slides: 10

Prokaryote Taxonomy & Diversity Classification, Nomenclature & Identification Phenetic Classification Molecular Phylogeny Approach • Classification (hierarchical grouping of taxa, based on characteristics) • Nomenclature (formal naming of taxa) • Identification (define characteristics that match a particular taxa). • Phylogeny (study of evolutionary relationships) • Bergey’s Manual of Systematic Bacteriology 1 st ed. (1984); mostly phenetic classification; 4 volumes 2 nd ed. (in prep); mostly phylogenetic classification; 5 volumes

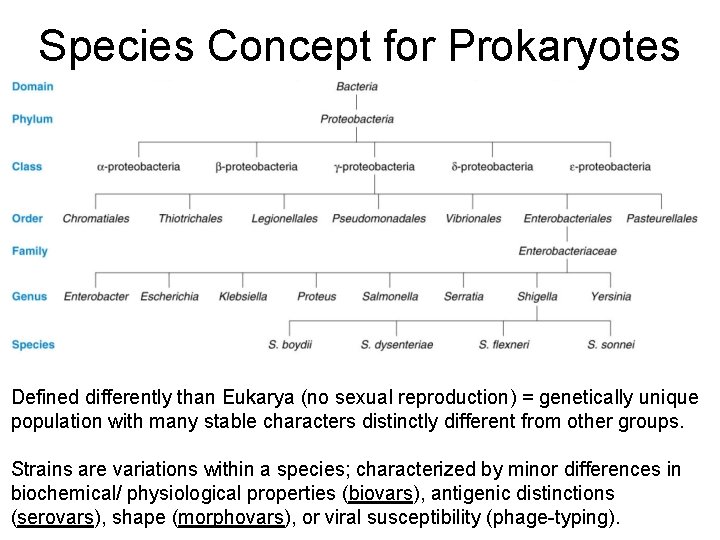
Species Concept for Prokaryotes Defined differently than Eukarya (no sexual reproduction) = genetically unique population with many stable characters distinctly different from other groups. Strains are variations within a species; characterized by minor differences in biochemical/ physiological properties (biovars), antigenic distinctions (serovars), shape (morphovars), or viral susceptibility (phage-typing).

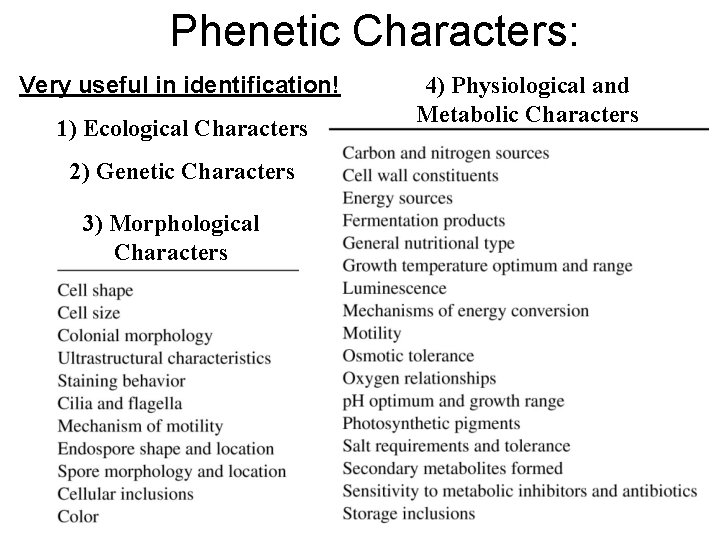
Phenetic Characters: Very useful in identification! 1) Ecological Characters 2) Genetic Characters 3) Morphological Characters 4) Physiological and Metabolic Characters

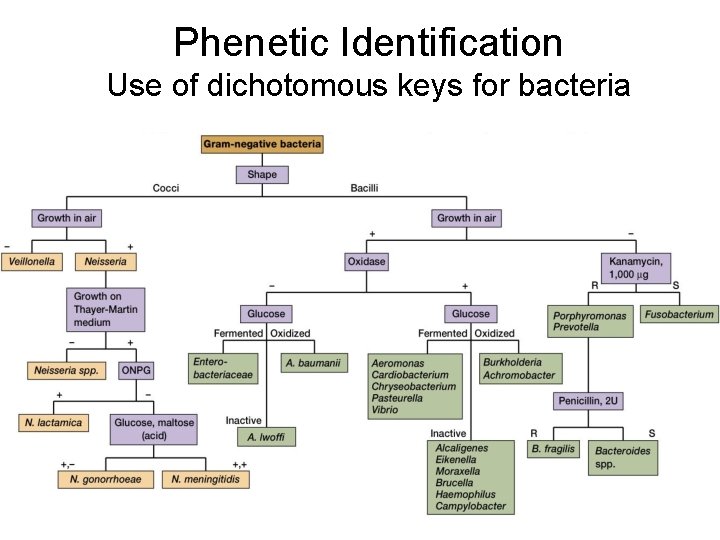
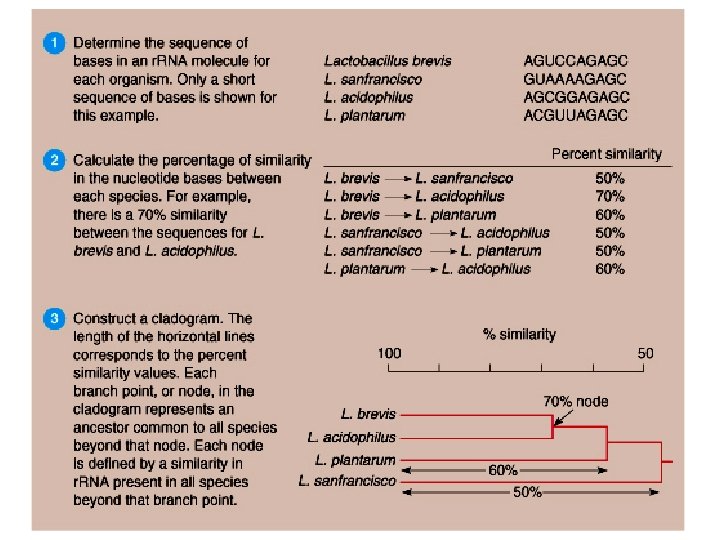
Phenetic Identification Use of dichotomous keys for bacteria

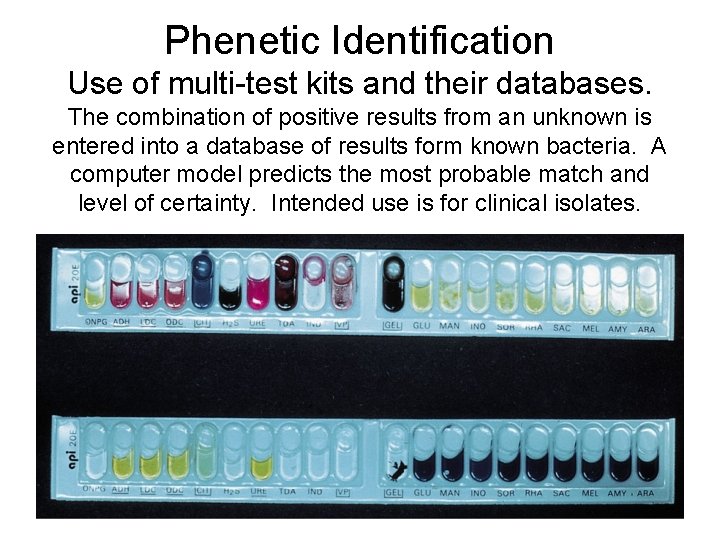
Phenetic Identification Use of multi-test kits and their databases. The combination of positive results from an unknown is entered into a database of results form known bacteria. A computer model predicts the most probable match and level of certainty. Intended use is for clinical isolates.

Molecular Characters • Fatty acid profiles (FAME analysis) • Proteins – Electrophoretic Mobility – Immuno-Reactivity – A. A. Sequence Data • Nucleic Acids – Nucleotide composition (G+C content ≈ Tm) – Degree of Hybridization (>70% ≈ species) – Nucleotide Sequence Data

Phylogenetic Classification Molecular Chronometers • Phylogeny refers to grouping based on evolutionary relatedness; regardless of phenetic characters. • Phylogeny is inferred from changes in protein or r. RNA sequence over time. • Attributes of an Ideal “Molecular Chronometer”: – – Universally distributed. Functionally homologous. Ease of analysis. Rate of sequence change commensurate with evolutionary distance measured.

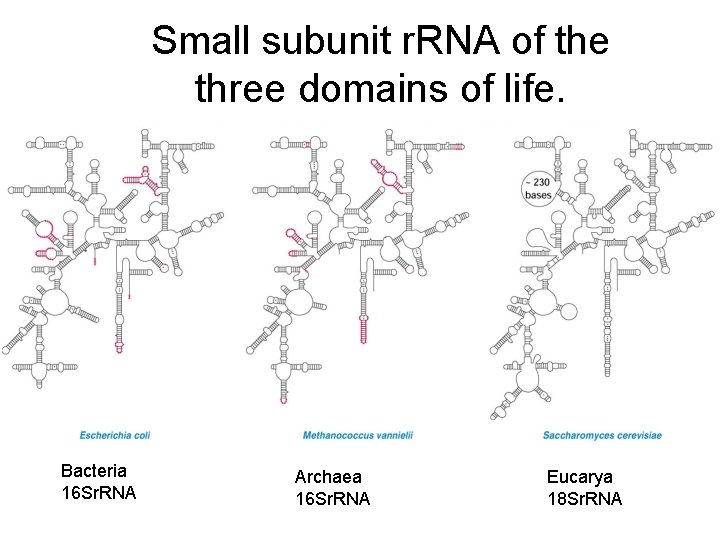
Small subunit r. RNA of the three domains of life. Bacteria 16 Sr. RNA Archaea 16 Sr. RNA Eucarya 18 Sr. RNA


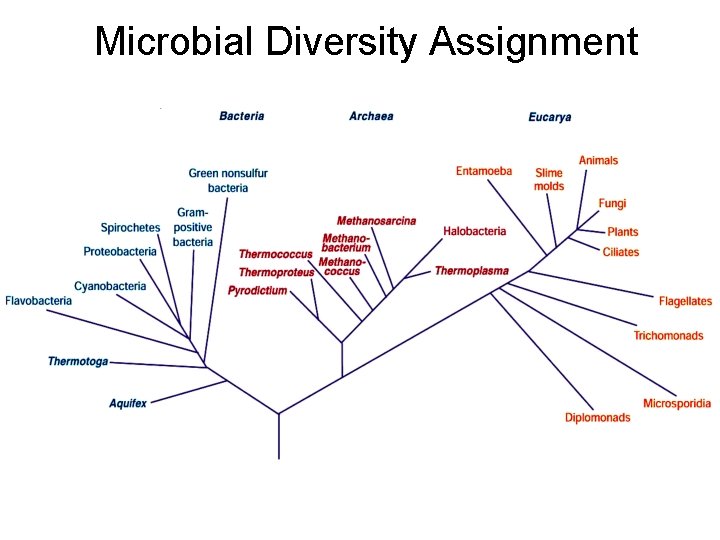
Microbial Diversity Assignment