Outline Randomization issues Twosample ttest vs paired ttest


![RGList class > attributes(Limmadata. C) $names [1] "R" "G" "Rb" "Gb" "weights" "targets" "genes" RGList class > attributes(Limmadata. C) $names [1] "R" "G" "Rb" "Gb" "weights" "targets" "genes"](https://slidetodoc.com/presentation_image_h2/d223c439aadc52844f4c14ddeed379a3/image-6.jpg)
![RGList class > Limmadata. C$R[1: 3, ] 51 -C 1 -3 -vs-W 1 -5 RGList class > Limmadata. C$R[1: 3, ] 51 -C 1 -3 -vs-W 1 -5](https://slidetodoc.com/presentation_image_h2/d223c439aadc52844f4c14ddeed379a3/image-7.jpg)
![RGList class Limmadata. C$genes$Status<-control. Status(spottypes, Limmadata. C) Limmadata. C$weights[Limmadata. C$genes$ID=="Blank"]<-0 Limmadata. C$printer<-get. Layout(Limmadata. C$genes) RGList class Limmadata. C$genes$Status<-control. Status(spottypes, Limmadata. C) Limmadata. C$weights[Limmadata. C$genes$ID=="Blank"]<-0 Limmadata. C$printer<-get. Layout(Limmadata. C$genes)](https://slidetodoc.com/presentation_image_h2/d223c439aadc52844f4c14ddeed379a3/image-8.jpg)


![limma > NBLimmadata. C<-background. Correct(Limmadata. C, method="none") > attributes(NBLimmadata. C) $names [1] "R" "G" limma > NBLimmadata. C<-background. Correct(Limmadata. C, method="none") > attributes(NBLimmadata. C) $names [1] "R" "G"](https://slidetodoc.com/presentation_image_h2/d223c439aadc52844f4c14ddeed379a3/image-11.jpg)





![Paired t-test using limma > mean(c(1, -1, 1, -1)*NNBLimmadata. C$M[2, ]) [1] -0. 03068036 Paired t-test using limma > mean(c(1, -1, 1, -1)*NNBLimmadata. C$M[2, ]) [1] -0. 03068036](https://slidetodoc.com/presentation_image_h2/d223c439aadc52844f4c14ddeed379a3/image-20.jpg)
![Paired t-test using limma > mean(c(1, -1, 1, -1)*NNBLimmadata. C$M[1, ]) [1] 0. 1425021 Paired t-test using limma > mean(c(1, -1, 1, -1)*NNBLimmadata. C$M[1, ]) [1] 0. 1425021](https://slidetodoc.com/presentation_image_h2/d223c439aadc52844f4c14ddeed379a3/image-21.jpg)

- Slides: 23

Outline • Randomization issues • Two-sample t-test vs paired t-test • I made a mistake in creating the dataset, so previous analyses will not be comparable with new ones • Issues of the background subtraction • Limma as the general tool for analyzing microarray data 1 -18 -2005 1

limma. . . is a package for the analysis of microarray data, especially the use of linear models for analyzing designed experiments and the assessment of differential expression. • Specially constructed data objects to represent various aspects of microarray data • Specially constructed "object methods" for importing, normalizing, displaying and analyzing microarray data • All objects and methods are transparent • All objects can be accessed and modified outside of limma • Unique in the implementation of the empirical Bayes procedure for identifying differentially expressed genes by "borrowing" information from different genes (everything so far has been gene by gene) 1 -18 -2005 2

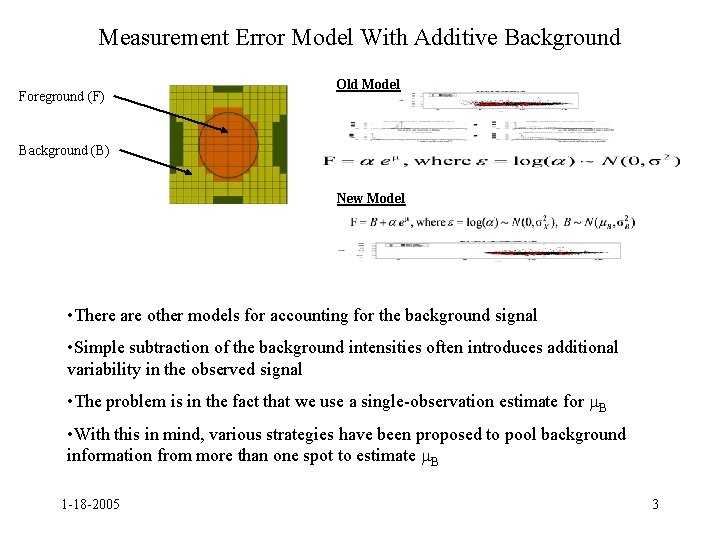
Measurement Error Model With Additive Background Foreground (F) Old Model Background (B) New Model • There are other models for accounting for the background signal • Simple subtraction of the background intensities often introduces additional variability in the observed signal • The problem is in the fact that we use a single-observation estimate for B • With this in mind, various strategies have been proposed to pool background information from more than one spot to estimate B 1 -18 -2005 3

limma Data to import: http: //eh 3. uc. edu/data/51 -C 1 -3 -vs-W 1 -5. gpr File descriptions: http: //eh 3. uc. edu/data/WTargets. txt Spot descriptions: http: //eh 3. uc. edu/data/WSpot. Types. txt Importing data: source("http: //eh 3. uc. edu/Limma. Data. Import. R") 1 -18 -2005 4

limma library(limma) data. directory<-"http: //eh 3. uc. edu/data/" targets<-read. Targets("http: //eh 3. uc. edu/data/WTargets. txt") spottypes<-read. Spot. Types("http: //eh 3. uc. edu/data/WSpot. Types. txt") Limmadata. C<-read. maimages(files=targets$File. Name, source="genepix", path = data. directory, columns=list(Gf = "F 532 Median", Gb ="B 532 Median", Rf = "F 635 Median", Rb = "B 635 Median"), annotation=c("Name", "ID", "Block", "Row", "Column"), wt. fun=wtflags(0)) 1 -18 -2005 5
![RGList class attributesLimmadata C names 1 R G Rb Gb weights targets genes RGList class > attributes(Limmadata. C) $names [1] "R" "G" "Rb" "Gb" "weights" "targets" "genes"](https://slidetodoc.com/presentation_image_h2/d223c439aadc52844f4c14ddeed379a3/image-6.jpg)
RGList class > attributes(Limmadata. C) $names [1] "R" "G" "Rb" "Gb" "weights" "targets" "genes" $class [1] "RGList" attr(, "package") [1] "limma" 1 -18 -2005 6
![RGList class Limmadata CR1 3 51 C 1 3 vsW 1 5 RGList class > Limmadata. C$R[1: 3, ] 51 -C 1 -3 -vs-W 1 -5](https://slidetodoc.com/presentation_image_h2/d223c439aadc52844f4c14ddeed379a3/image-7.jpg)
RGList class > Limmadata. C$R[1: 3, ] 51 -C 1 -3 -vs-W 1 -5 60 -W 2 -3 -vs-C 2 -5 72 -C 3 -3 -vs-W 3 -5 79 -W 4 -3 -vs-C 4 -5 82 -C 5 -3 -vs-W 5 -5 97 -W 6 -3 -vs-C 6 -5 [1, ] 85 57 91 71 [2, ] 358 1102 2394 1685 [3, ] 168 376 620 67 111 575 882 206 293 > Limmadata. C$Rb[1: 3, ] 51 -C 1 -3 -vs-W 1 -5 60 -W 2 -3 -vs-C 2 -5 72 -C 3 -3 -vs-W 3 -5 79 -W 4 -3 -vs-C 4 -5 82 -C 5 -3 -vs-W 5 -5 97 -W 6 -3 -vs-C 6 -5 [1, ] 81 55 65 51 52 72 [2, ] 81 56 65 51 51 72 [3, ] 82 57 64 50 48 69 > Limmadata. C$genes[1, ] Name ID Block Row Column 1 no name Rn 30000100 1 -18 -2005 1 1 1 7
![RGList class Limmadata CgenesStatuscontrol Statusspottypes Limmadata C Limmadata CweightsLimmadata CgenesIDBlank0 Limmadata Cprinterget LayoutLimmadata Cgenes RGList class Limmadata. C$genes$Status<-control. Status(spottypes, Limmadata. C) Limmadata. C$weights[Limmadata. C$genes$ID=="Blank"]<-0 Limmadata. C$printer<-get. Layout(Limmadata. C$genes)](https://slidetodoc.com/presentation_image_h2/d223c439aadc52844f4c14ddeed379a3/image-8.jpg)
RGList class Limmadata. C$genes$Status<-control. Status(spottypes, Limmadata. C) Limmadata. C$weights[Limmadata. C$genes$ID=="Blank"]<-0 Limmadata. C$printer<-get. Layout(Limmadata. C$genes) > attributes(Limmadata. C) $names [1] "R" "G" "Rb" "Gb" "weights" "targets" "genes" "printer" $class [1] "RGList" attr(, "package") [1] "limma" > Limmadata. C$genes[1, ] Name ID Block Row Column Status 1 no name Rn 30000100 1 -18 -2005 1 1 1 c. DNA 8

Plotting data in a RGList object > plot. MA(Limmadata. C, array=1, xlim=c(-1, 16), ylim=c(-3, 8)) 1 -18 -2005 9

limma • Plot. MA automatically subtracts the background intensities before plotting data • Does not plot data with weight 0 • If you want to plot all data or the data without subtracting background, you need to do a little work • source("http: //eh 3. uc. edu/Background. Effects. R") 1 -18 -2005 10
![limma NBLimmadata Cbackground CorrectLimmadata C methodnone attributesNBLimmadata C names 1 R G limma > NBLimmadata. C<-background. Correct(Limmadata. C, method="none") > attributes(NBLimmadata. C) $names [1] "R" "G"](https://slidetodoc.com/presentation_image_h2/d223c439aadc52844f4c14ddeed379a3/image-11.jpg)
limma > NBLimmadata. C<-background. Correct(Limmadata. C, method="none") > attributes(NBLimmadata. C) $names [1] "R" "G" "weights" "targets" "genes" "printer" $class [1] "RGList" attr(, "package") [1] "limma" • Note that background measurements are gone 1 -18 -2005 11

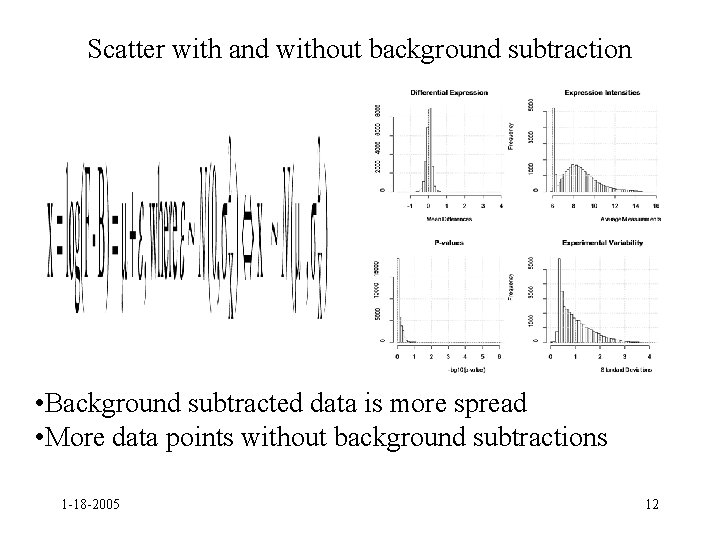
Scatter with and without background subtraction • Background subtracted data is more spread • More data points without background subtractions 1 -18 -2005 12

Plotting all data points • Want to plot data points with weight 0 as well • Create datasets with and without subtracting background and set all weights to 1 Spots. Per. Array<-dim(Limmadata. C$R)[1] Narrays<-dim(Limmadata. C$R)[2] Limmadata<-Limmadata. C Limmadata$weights[1: Spots. Per. Array, 1: Narrays]<-1 NBLimmadata<-NBLimmadata. C NBLimmadata$weights[1: Spots. Per. Array, 1: Narrays]<-1 1 -18 -2005 13

Plotting all data points All data Zero-weight data removed Background Subtracted Raw 1 -18 -2005 14

Which one to use? • Removing points with the weight zero seems reasonable • Subtracting background costs us some data points even if one channel is above background since differences of log-transformed measurements are used only • Subtracting background seems to increase the variability, but it is unclear how would this affect results • For now proceed without background subtraction, but compare results at the end • Exploring other proposed background-adjustment methods also seems like a good idea 1 -18 -2005 15

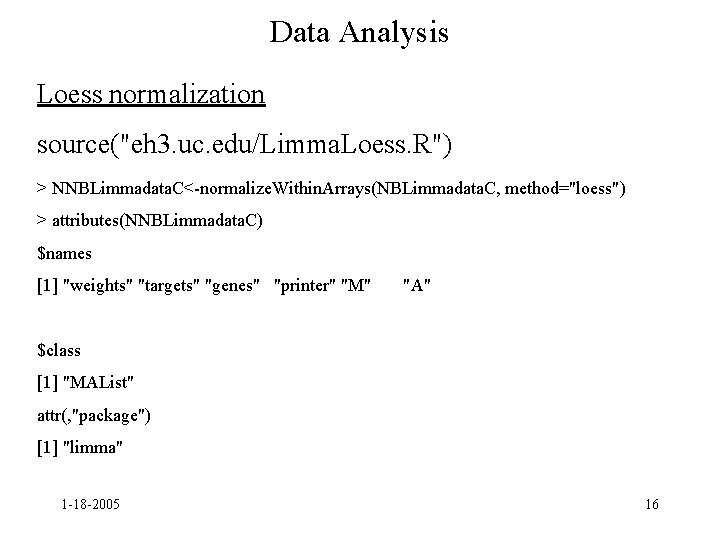
Data Analysis Loess normalization source("eh 3. uc. edu/Limma. Loess. R") > NNBLimmadata. C<-normalize. Within. Arrays(NBLimmadata. C, method="loess") > attributes(NNBLimmadata. C) $names [1] "weights" "targets" "genes" "printer" "M" "A" $class [1] "MAList" attr(, "package") [1] "limma" 1 -18 -2005 16

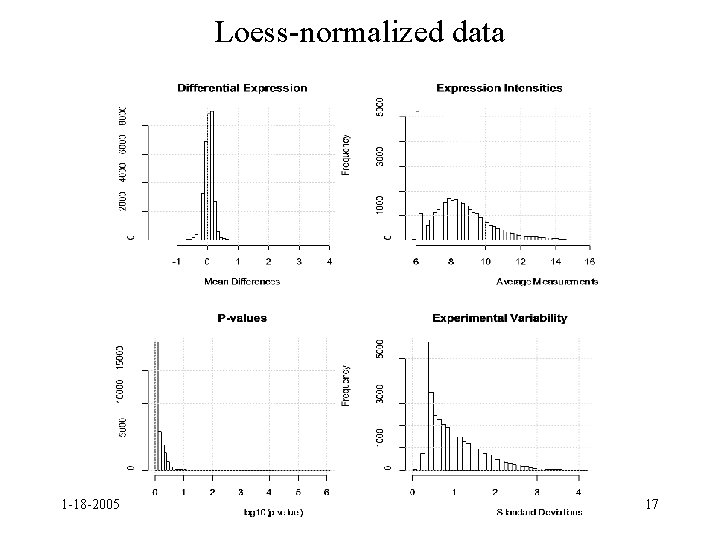
Loess-normalized data 1 -18 -2005 17

Paired t-test using limma source("http: //eh 3. uc. edu/Limma. TTest. R") > design<-model. Matrix(targets, ref="C") Found unique target names: CW > design W 51 -C 1 -3 -vs-W 1 -5 1 60 -W 2 -3 -vs-C 2 -5 -1 72 -C 3 -3 -vs-W 3 -5 1 79 -W 4 -3 -vs-C 4 -5 -1 82 -C 5 -3 -vs-W 5 -5 1 97 -W 6 -3 -vs-C 6 -5 -1 1 -18 -2005 18

Paired t-test using limma > Limma. PTT<-lm. Fit(MA, design) Error in. class 1(object) : Object "MA" not found > Limma. PTT<-lm. Fit(NNBLimmadata. C, design) > > attributes(Limma. PTT) $names [1] "coefficients" "stdev. unscaled" "sigma" [5] "cov. coefficients" "pivot" [9] "genes" "method" "df. residual" "design" "Amean" $class [1] "MArray. LM" attr(, "package") [1] "limma" 1 -18 -2005 19
![Paired ttest using limma meanc1 1 1 1NNBLimmadata CM2 1 0 03068036 Paired t-test using limma > mean(c(1, -1, 1, -1)*NNBLimmadata. C$M[2, ]) [1] -0. 03068036](https://slidetodoc.com/presentation_image_h2/d223c439aadc52844f4c14ddeed379a3/image-20.jpg)
Paired t-test using limma > mean(c(1, -1, 1, -1)*NNBLimmadata. C$M[2, ]) [1] -0. 03068036 > var(c(1, -1, 1, -1)*NNBLimmadata. C$M[2, ])^0. 5 [1] 0. 3176068 > 1/(6^0. 5) [1] 0. 4082483 > mean(c(1, -1, 1, -1)*NNBLimmadata. C$M[2, ])/((var(c(1, -1, 1, -1)*NNBLimmadata. C$M[2, ])^0. 5)*(1/(6^0. 5))) [1] -0. 2366172 > > Limma. PTT$coefficients[2] [1] -0. 03068036 > Limma. PTT$stdev. unscaled[2] [1] 0. 4082483 > Limma. PTT$sigma[2] [1] 0. 3176068 > Limma. PTT$coefficients[2]/(Limma. PTT$sigma[2]*Limma. PTT$stdev. unscaled[2]) [1] -0. 2366172 1 -18 -2005 20
![Paired ttest using limma meanc1 1 1 1NNBLimmadata CM1 1 0 1425021 Paired t-test using limma > mean(c(1, -1, 1, -1)*NNBLimmadata. C$M[1, ]) [1] 0. 1425021](https://slidetodoc.com/presentation_image_h2/d223c439aadc52844f4c14ddeed379a3/image-21.jpg)
Paired t-test using limma > mean(c(1, -1, 1, -1)*NNBLimmadata. C$M[1, ]) [1] 0. 1425021 > var(c(1, -1, 1, -1)*NNBLimmadata. C$M[1, ])^0. 5 [1] 0. 2395690 > 1/(6^0. 5) [1] 0. 4082483 > mean(c(1, -1, 1, -1)*NNBLimmadata. C$M[1, ])/((var(c(1, -1, 1, -1)*NNBLimmadata. C$M[1, ])^0. 5)*(1/(6^0. 5))) [1] 1. 457023 > > Limma. PTT$coefficients[1] 0. 1875361 > Limma. PTT$stdev. unscaled[1] 0. 5 > Limma. PTT$sigma[1] 0. 2831248 > Limma. PTT$coefficients[1]/(Limma. PTT$sigma[1]*Limma. PTT$stdev. unscaled[1]) [1] 1. 324760 > NNBLimmadata. C$weights[1, ] 51 -C 1 -3 -vs-W 1 -5 60 -W 2 -3 -vs-C 2 -5 72 -C 3 -3 -vs-W 3 -5 79 -W 4 -3 -vs-C 4 -5 82 -C 5 -3 -vs-W 5 -5 97 -W 6 -3 -vs-C 6 -5 0 1 -18 -2005 0 1 1 21

Paired t-test using limma > dfp<-Limma. PTT$df. residuals>0 > Limma. PTT$Limma. TStat<-Limma. PTT$coefficients/(Limma. PTT$sigma*Limma. PTT$stdev. unscaled) > Limma. PTT$Limma. TPvalue<-rep(NA, Spots. Per. Array) > Limma. PTT$Limma. TPvalue[dfp]<-2*pt(Limma. PTT$Limma. TStat[dfp], Limma. PTT$df. residual[dfp], lower. tail =FALSE) > attributes(Limma. PTT) $names [1] "coefficients" "stdev. unscaled" "sigma" [5] "cov. coefficients" "pivot" [9] "genes" "Amean" "method" "Limma. TStat" "df. residual" "design" "Limma. TPvalue" $class [1] "MArray. LM" attr(, "package") [1] "limma" 1 -18 -2005 22

limma so far • Facilitates easy data import and normalization • Keeps track of "bad" spots • To run the basic t-test, it takes a bit of additional work • If we were to use the empirical Bayes statistics as implemented in limma, it would be even easier • Empirical Bayes is generally BETTER than simple t-test • Will talk about this type of analysis next week • Limma also allows fitting models with multiple factors which we will also talk about next week • Next time – multiple hypothesis testing and p-value adjustments 1 -18 -2005 23