Meccanismi di riparazione Donata Orioli Risposta cellulare al





- Slides: 20


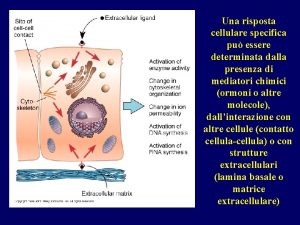
Meccanismi di riparazione Donata Orioli Risposta cellulare al danno Ivana Scovassi RNA Giuseppe Biamonti

Difetti nei meccanismi di riparazione del DNA e malattie associate Donata Orioli/Giovanni Maga 29 Febbraio 2016 Riunione CNAO-CNR-INFN

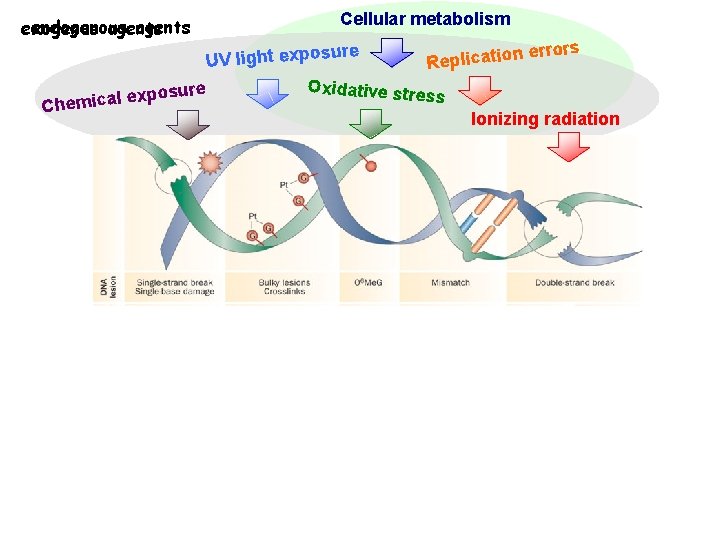
DNA damaging agents DNA Repair systems

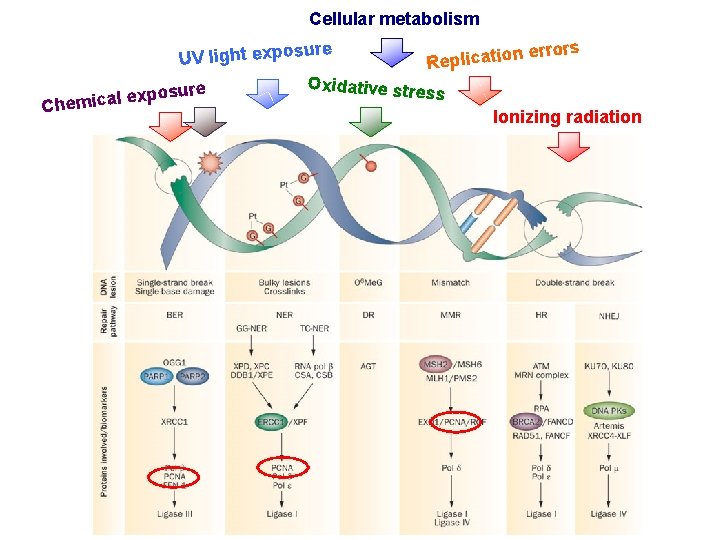
endogenous agents exogenus agents Cellular metabolism re n errors o i t a c i l p UV light exposu e R Oxidative stre osure p x ss e l a c i Chem Ionizing radiation

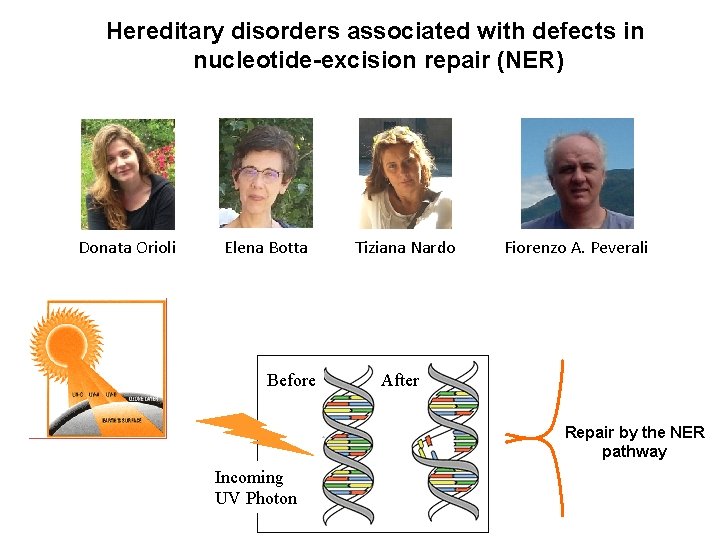
Hereditary disorders associated with defects in nucleotide-excision repair (NER) Donata Orioli Elena Botta Before Tiziana Nardo Fiorenzo A. Peverali After Repair by the NER pathway Incoming UV Photon

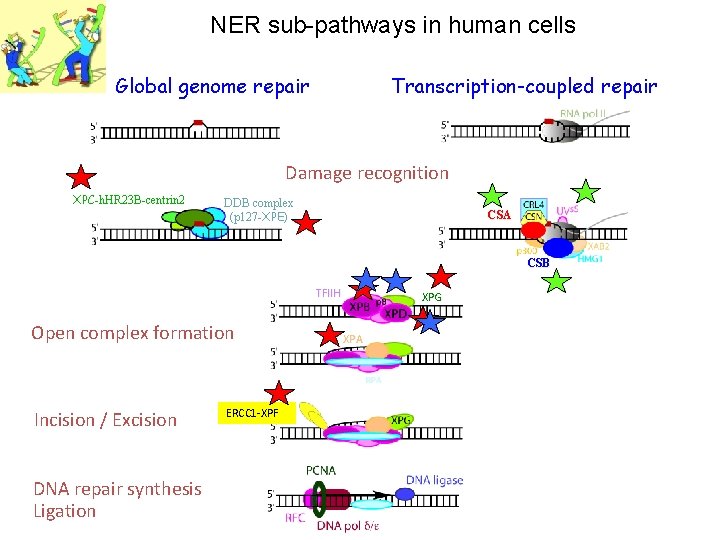
NER sub-pathways in human cells Global genome repair Transcription-coupled repair Damage recognition XPC-h. HR 23 B-centrin 2 DDB complex (p 127 -XPE) CSA CSB TFIIH Open complex formation XPG XPA RPA Incision / Excision DNA repair synthesis Ligation ERCC 1 -XPF XPG

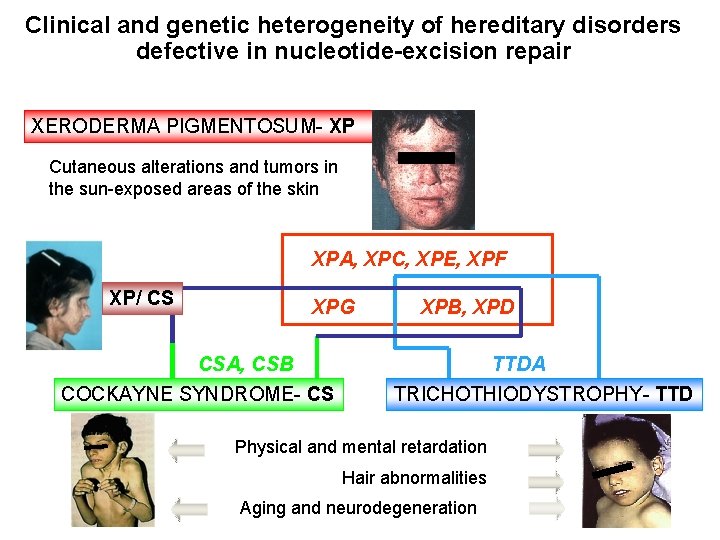
Clinical and genetic heterogeneity of hereditary disorders defective in nucleotide-excision repair XERODERMA PIGMENTOSUM- XP Cutaneous alterations and tumors in the sun-exposed areas of the skin XPA, XPC, XPE, XPF XP/ CS XPG CSA, CSB COCKAYNE SYNDROME- CS XPB, XPD TTDA TRICHOTHIODYSTROPHY- TTD Physical and mental retardation Hair abnormalities Aging and neurodegeneration

Collection of patient material • In vitro establishment of cell strains and lines from patients and their relatives; • Evaluation of the DNA repair defect; • Identification of the gene/mutations responsible for the pathological phenotype. • to confirm clinical diagnosis; • to collect new material from homozygous and heterozygous carriers with NER defects; • to identify new NER genes; • to establish the relationship between clinical symptoms and molecular defects

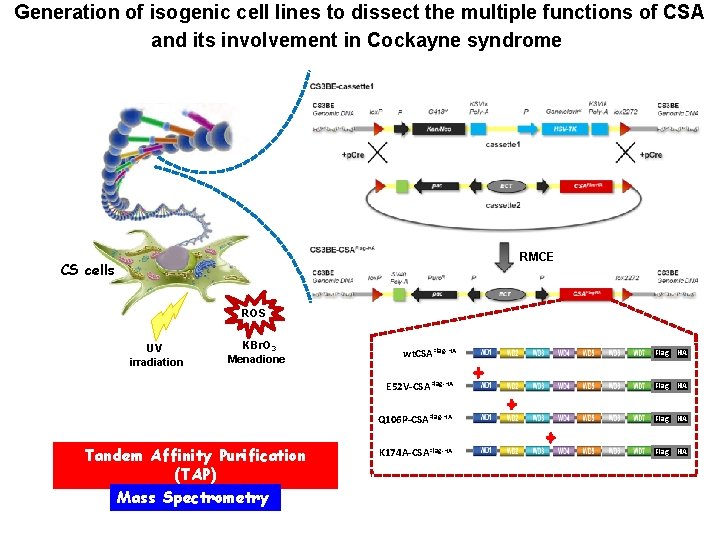
Generation of isogenic cell lines to dissect the multiple functions of CSA and its involvement in Cockayne syndrome RMCE CS cells ROS UV irradiation KBr. O 3 Menadione Tandem Affinity Purification (TAP) Mass Spectrometry wt. CSAFlag-HA Flag HA E 52 V-CSAFlag-HA Flag HA Q 106 P-CSAFlag-HA Flag HA K 174 A-CSAFlag-HA Flag HA

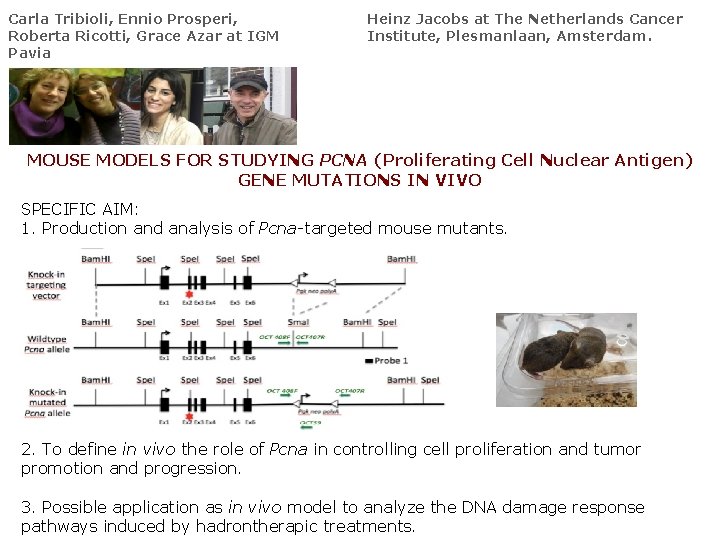
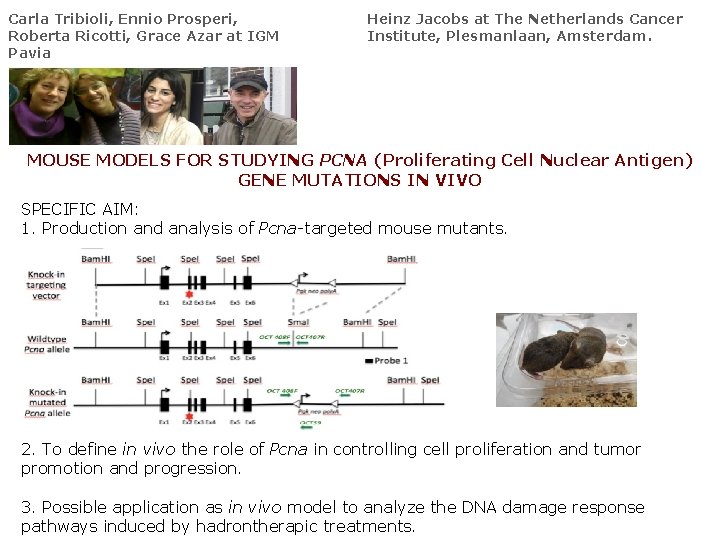
Carla Tribioli, Ennio Prosperi, Roberta Ricotti, Grace Azar at IGM Pavia Heinz Jacobs at The Netherlands Cancer Institute, Plesmanlaan, Amsterdam. MOUSE MODELS FOR STUDYING PCNA (Proliferating Cell Nuclear Antigen) GENE MUTATIONS IN VIVO SPECIFIC AIM: 1. Production and analysis of Pcna-targeted mouse mutants. 2. To define in vivo the role of Pcna in controlling cell proliferation and tumor promotion and progression. 3. Possible application as in vivo model to analyze the DNA damage response pathways induced by hadrontherapic treatments.

Cellular metabolism re n errors o i t a c UV light exposu i l p e R Oxidative stre e r u s ss o p x e l a c i m e h C Ionizing radiation

Carla Tribioli, Ennio Prosperi, Roberta Ricotti, Grace Azar at IGM Pavia Heinz Jacobs at The Netherlands Cancer Institute, Plesmanlaan, Amsterdam. MOUSE MODELS FOR STUDYING PCNA (Proliferating Cell Nuclear Antigen) GENE MUTATIONS IN VIVO SPECIFIC AIM: 1. Production and analysis of Pcna-targeted mouse mutants. 2. To define in vivo the role of Pcna in controlling cell proliferation and tumor promotion and progression. 3. Possible application as in vivo model to analyze the DNA damage response pathways induced by hadrontherapic treatments.

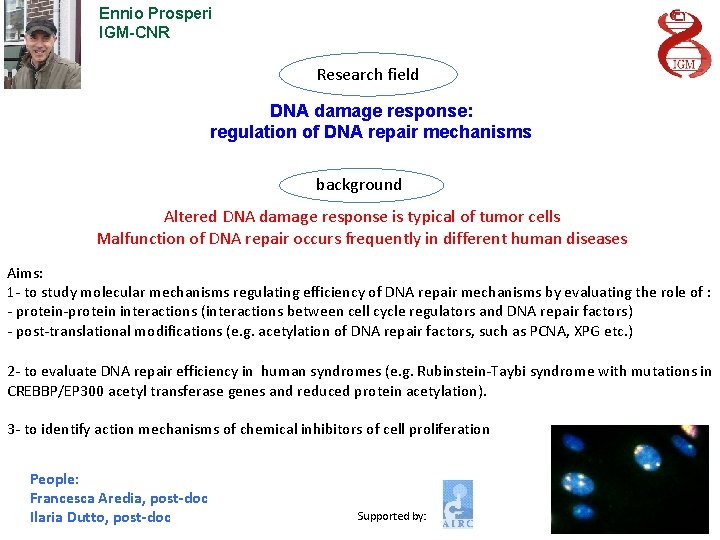
Ennio Prosperi IGM-CNR Research field DNA damage response: regulation of DNA repair mechanisms background Altered DNA damage response is typical of tumor cells Malfunction of DNA repair occurs frequently in different human diseases Aims: 1 - to study molecular mechanisms regulating efficiency of DNA repair mechanisms by evaluating the role of : - protein-protein interactions (interactions between cell cycle regulators and DNA repair factors) - post-translational modifications (e. g. acetylation of DNA repair factors, such as PCNA, XPG etc. ) 2 - to evaluate DNA repair efficiency in human syndromes (e. g. Rubinstein-Taybi syndrome with mutations in CREBBP/EP 300 acetyl transferase genes and reduced protein acetylation). 3 - to identify action mechanisms of chemical inhibitors of cell proliferation People: Francesca Aredia, post-doc Ilaria Dutto, post-doc Supported by:

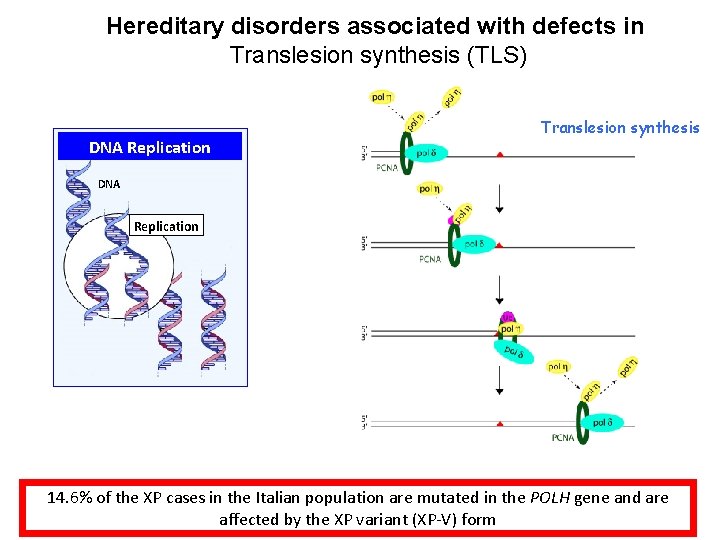
Hereditary disorders associated with defects in Translesion synthesis (TLS) DNA Replication Translesion synthesis DNA Replication 14. 6% of the XP cases in the Italian population are mutated in the POLH gene and are affected by the XP variant (XP-V) form

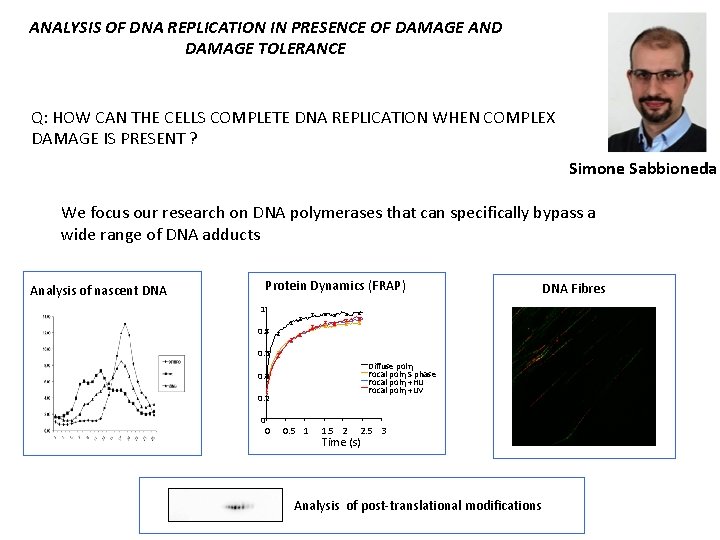
ANALYSIS OF DNA REPLICATION IN PRESENCE OF DAMAGE AND DAMAGE TOLERANCE Q: HOW CAN THE CELLS COMPLETE DNA REPLICATION WHEN COMPLEX DAMAGE IS PRESENT ? Simone Sabbioneda We focus our research on DNA polymerases that can specifically bypass a wide range of DNA adducts Analysis of nascent DNA Protein Dynamics (FRAP) 1 0. 8 0. 6 Diffuse polh Focal polh S phase Focal polh +HU Focal polh +UV 0. 4 0. 2 0 0 0. 5 1 1. 5 2 2. 5 3 Time (s) Analysis of post-translational modifications DNA Fibres

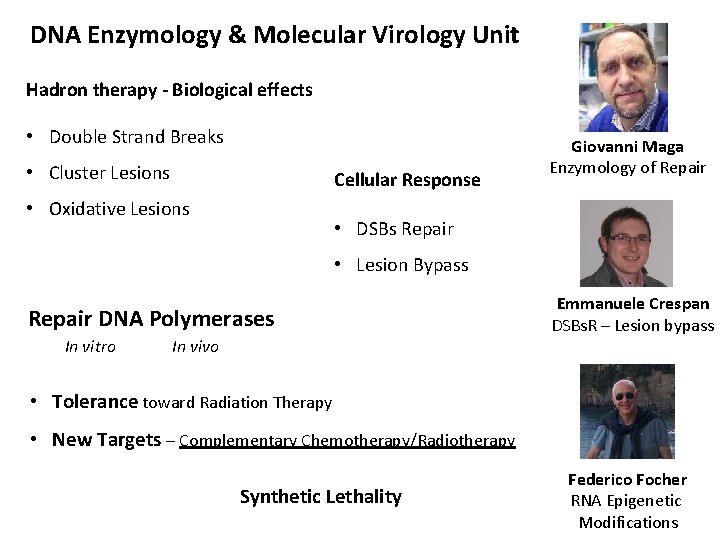
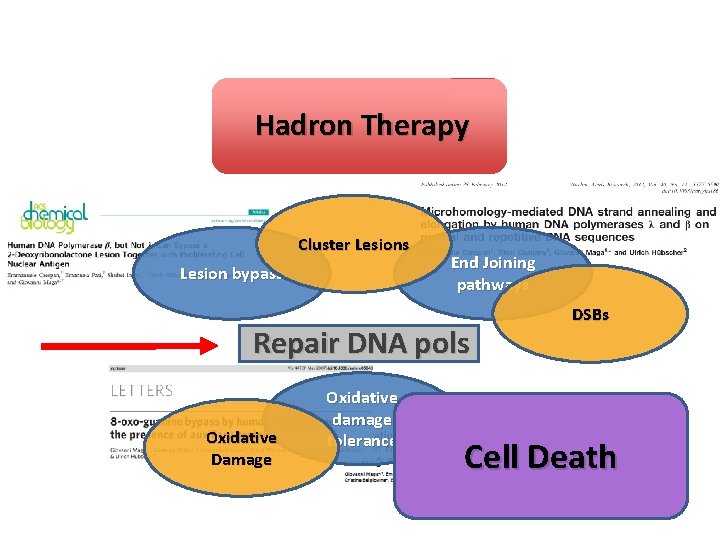
DNA Enzymology & Molecular Virology Unit Hadron therapy - Biological effects • Double Strand Breaks • Cluster Lesions Cellular Response • Oxidative Lesions Giovanni Maga Enzymology of Repair • DSBs Repair • Lesion Bypass Repair DNA Polymerases In vitro In vivo Emmanuele Crespan DSBs. R – Lesion bypass • Tolerance toward Radiation Therapy • New Targets – Complementary Chemotherapy/Radiotherapy Synthetic Lethality Federico Focher RNA Epigenetic Modifications

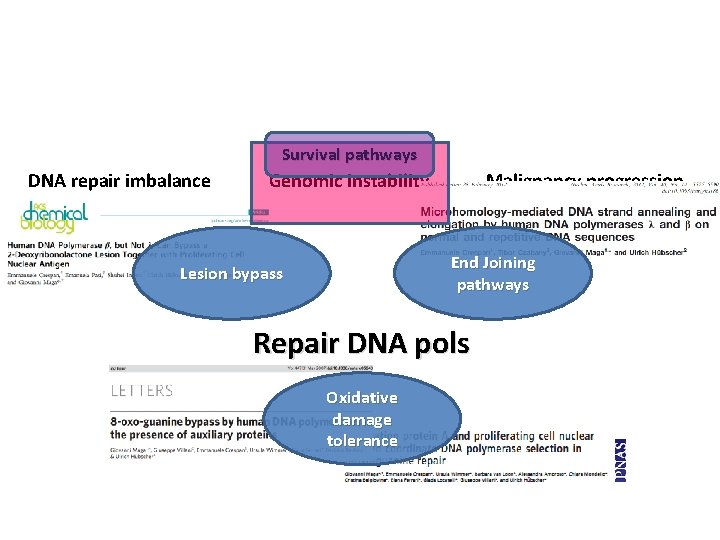
Survival pathways DNA repair imbalance Genomic instability Malignancy progression End Joining pathways Lesion bypass Repair DNA pols Oxidative damage tolerance

Hadron Therapy Cluster Lesions Lesion bypass End Joining pathways Repair DNA pols Oxidative Damage Oxidative damage tolerance DSBs Cell Death

 Donata orioli
Donata orioli Oitac
Oitac La regolazione degli affetti e la riparazione del sé
La regolazione degli affetti e la riparazione del sé Test di appercezione tematica tavole
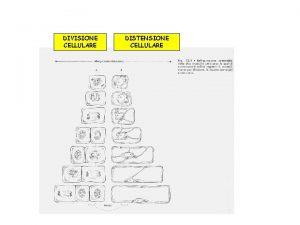
Test di appercezione tematica tavole Meccanismi energetici mappa concettuale
Meccanismi energetici mappa concettuale Meccanismi epigenetici
Meccanismi epigenetici Valvola di von gubaroff
Valvola di von gubaroff Meccanismi locali di ribaltamento
Meccanismi locali di ribaltamento 5 parti dell'organizzazione
5 parti dell'organizzazione Tvams
Tvams Maria podolak-dawidziak
Maria podolak-dawidziak Donata urbaniak kujda
Donata urbaniak kujda Donata mauricaitė
Donata mauricaitė Omino attento
Omino attento Stimolo chiuso risposta aperta
Stimolo chiuso risposta aperta Mostra link per inviare un'altra risposta
Mostra link per inviare un'altra risposta Modello stimolo-risposta marketing
Modello stimolo-risposta marketing Domande a risposta multipla
Domande a risposta multipla Domanda motivazionale tfa sostegno come rispondere
Domanda motivazionale tfa sostegno come rispondere Risposta
Risposta Risposta primaria e secondaria
Risposta primaria e secondaria