JS 190 Introduction to STRs Continued I Learning










- Slides: 30

JS 190 - Introduction to STRs- Continued I. Learning Objectives (C 6 Butler) a. Short Tandem Repeats 1. CE artifacts and Fluorescent Dye multiplexing revisited 2. Biology of STRs- Define. Balance Stutter Products Non-template Addition Microvariants Null Alleles Mutation Rates

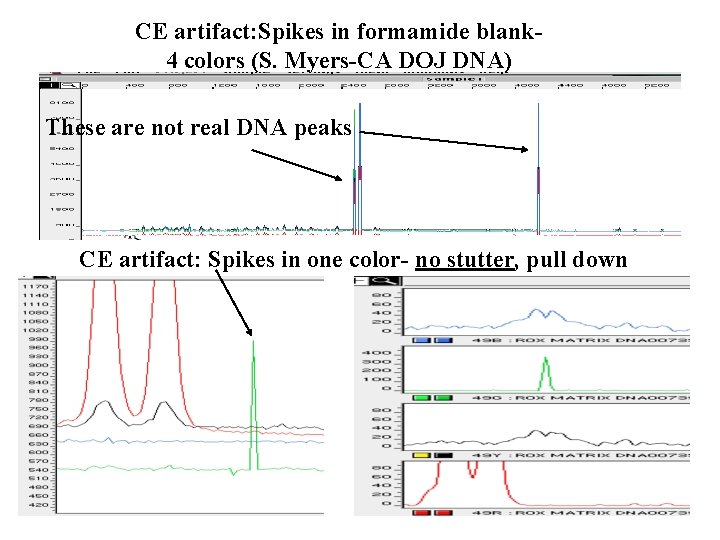
CE artifact: Spikes in formamide blank 4 colors (S. Myers-CA DOJ DNA) These are not real DNA peaks • CE artifact: Spikes in one color- no stutter, pull down

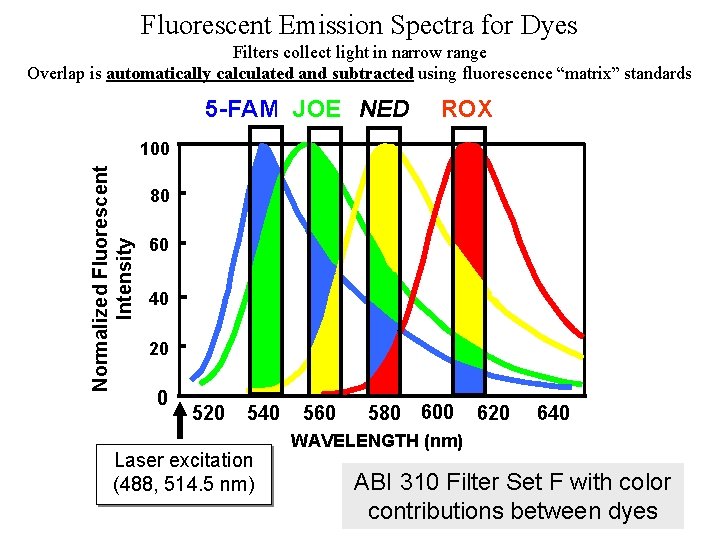
Fluorescent Emission Spectra for Dyes Filters collect light in narrow range Overlap is automatically calculated and subtracted using fluorescence “matrix” standards 5 -FAM JOE NED ROX Normalized Fluorescent Intensity 100 80 60 40 20 0 520 540 Laser excitation (488, 514. 5 nm) 560 580 600 620 640 WAVELENGTH (nm) ABI 310 Filter Set F with color contributions between dyes

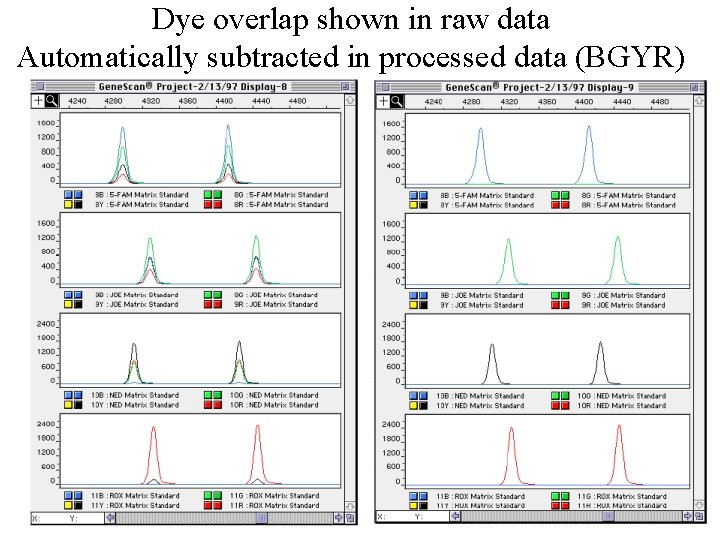
Dye overlap shown in raw data Automatically subtracted in processed data (BGYR)

Short Tandem Repeats: a subgroup of tandem repeats (Kuhl and Caskey 1993. Curr. Opin. in Genet. Dev. 3: 404) • Head to tail arrangements of sequence units (4 bp), • Common in genomes (thousands distributed) • Polymorphic: vary in length by no. of and/or by content of repeats. • Stably inherited on a human time scale (for most) • Well studied b/c others are implicated in Human Diseases and therefore the subject of clinical studies.

Trinucleotide Repeats Implicated in Human Diseases Sutherland Richards. 1994. Dynamic Mutations American Scientist 82: 157

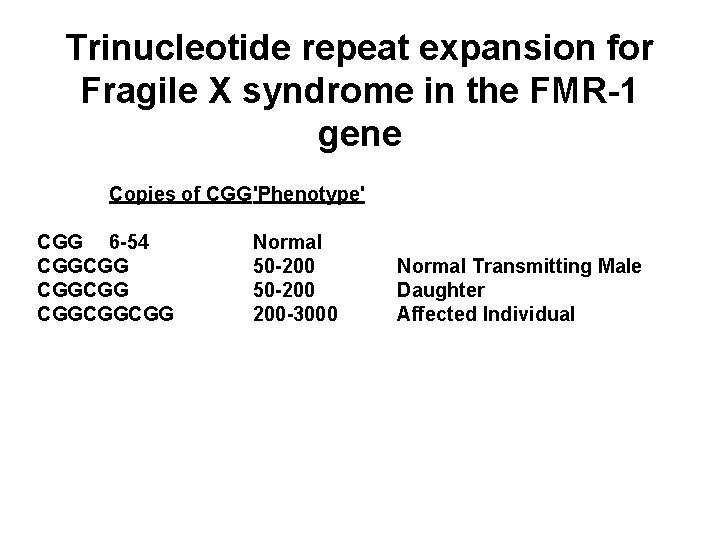
Trinucleotide repeat expansion for Fragile X syndrome in the FMR-1 gene Copies of CGG'Phenotype' CGG 6 -54 CGGCGGCGG Normal 50 -200 200 -3000 Normal Transmitting Male Daughter Affected Individual

Advantages of STRs in Forensics • All of the above and more! Common, polymorphic, stably inherited, well studied- discrete sizes • Small size and size range- Useful on highly degraded samples • Small size range- Less prone to preferential amplification of the smaller allele • Multiple STRs provide powerful discrimination • Abundance permits choice of STRs with non overlapping size ranges. • Even for those with overlapping sizes, use of different color fluorescently tagged primers permit rapid automated analysis.

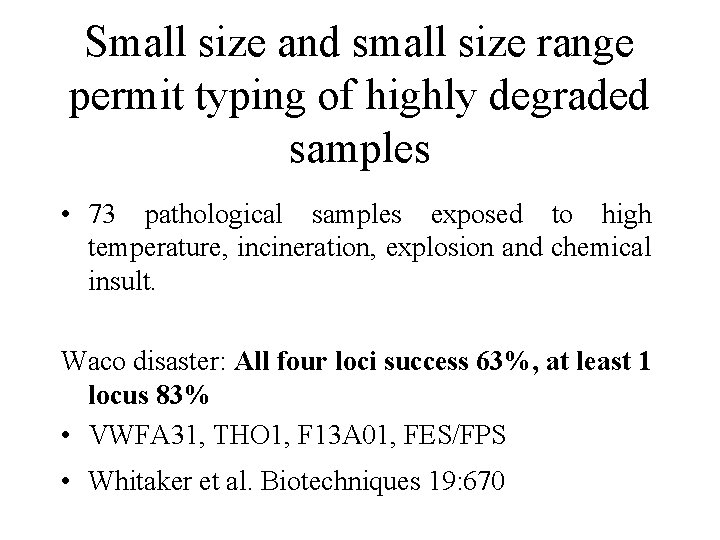
Small size and small size range permit typing of highly degraded samples • 73 pathological samples exposed to high temperature, incineration, explosion and chemical insult. Waco disaster: All four loci success 63%, at least 1 locus 83% • VWFA 31, THO 1, F 13 A 01, FES/FPS • Whitaker et al. Biotechniques 19: 670

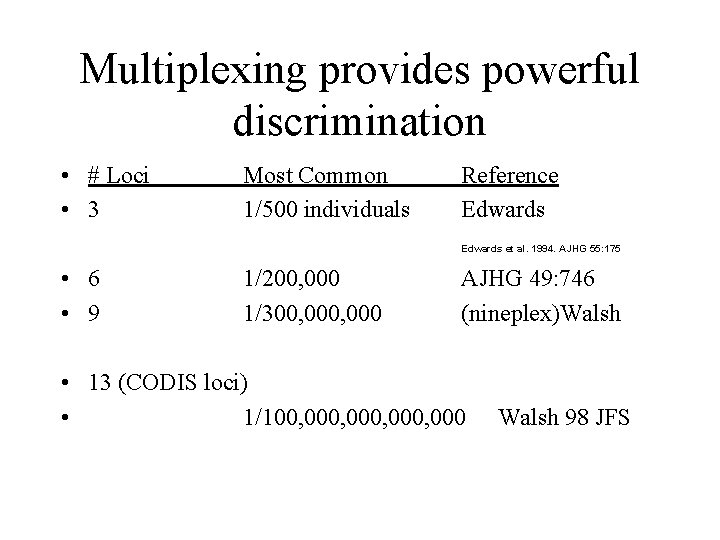
Multiplexing provides powerful discrimination • # Loci • 3 Most Common 1/500 individuals Reference Edwards et al. 1994. AJHG 55: 175 • 6 • 9 1/200, 000 1/300, 000 AJHG 49: 746 (nineplex)Walsh • 13 (CODIS loci) • 1/100, 000, 000 Walsh 98 JFS

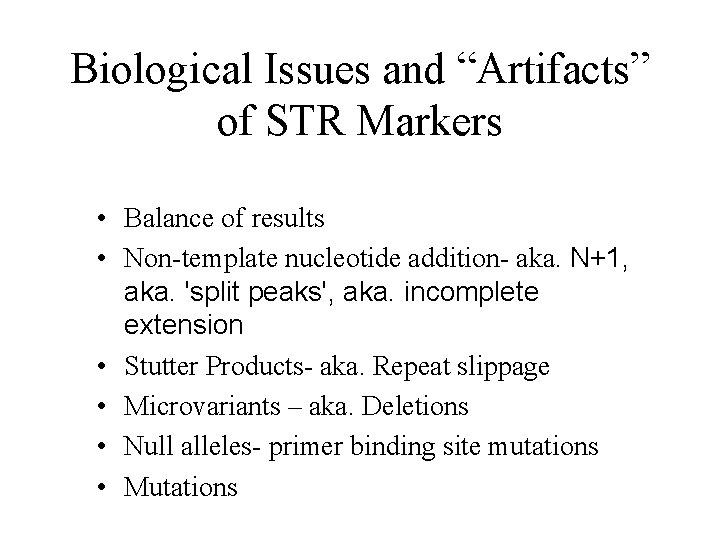
Biological Issues and “Artifacts” of STR Markers • Balance of results • Non-template nucleotide addition- aka. N+1, aka. 'split peaks', aka. incomplete extension • Stutter Products- aka. Repeat slippage • Microvariants – aka. Deletions • Null alleles- primer binding site mutations • Mutations

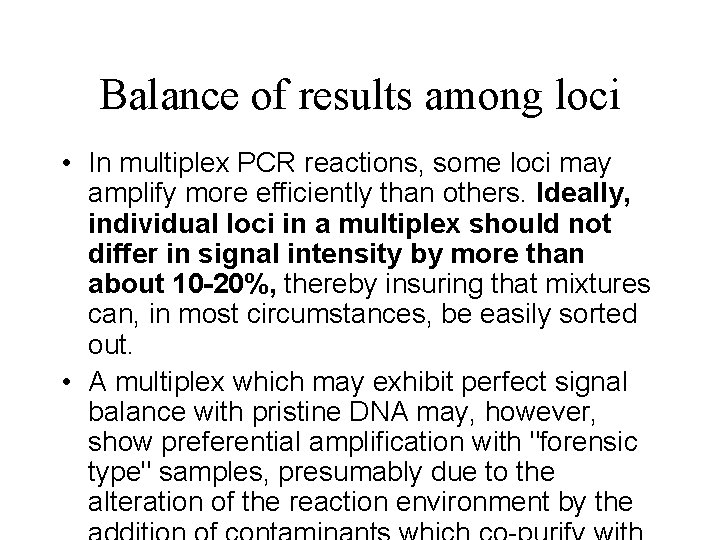
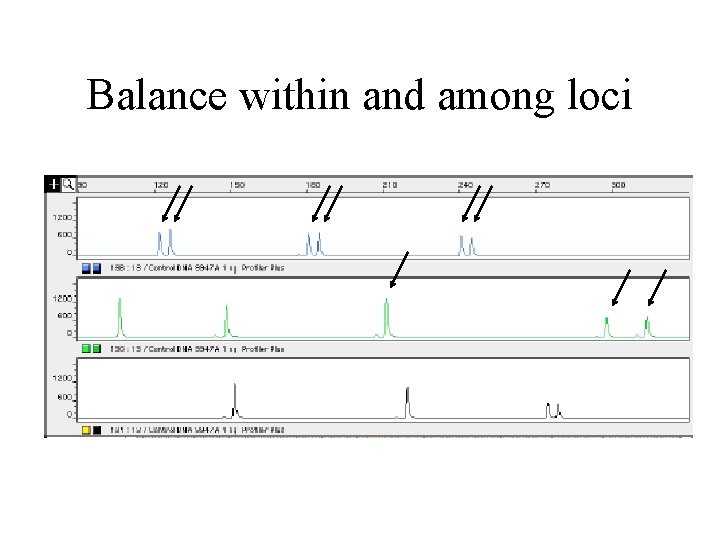
Balance of results among loci • In multiplex PCR reactions, some loci may amplify more efficiently than others. Ideally, individual loci in a multiplex should not differ in signal intensity by more than about 10 -20%, thereby insuring that mixtures can, in most circumstances, be easily sorted out. • A multiplex which may exhibit perfect signal balance with pristine DNA may, however, show preferential amplification with "forensic type" samples, presumably due to the alteration of the reaction environment by the

Balance within and among loci

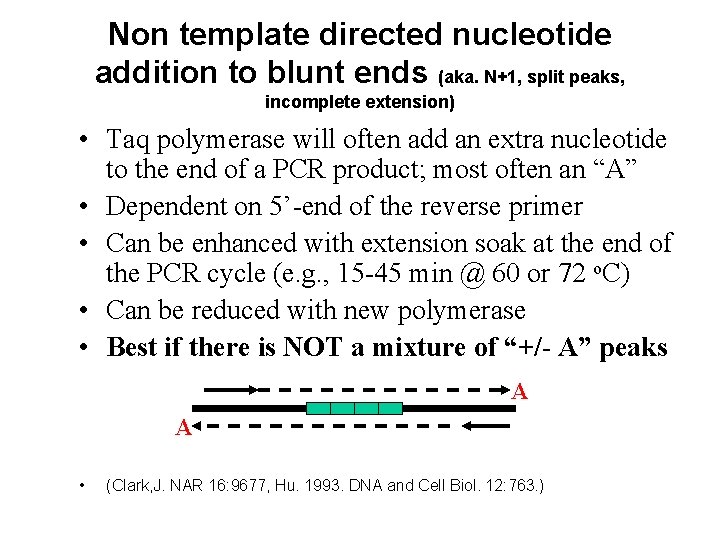
Non template directed nucleotide addition to blunt ends (aka. N+1, split peaks, incomplete extension) • Taq polymerase will often add an extra nucleotide to the end of a PCR product; most often an “A” • Dependent on 5’-end of the reverse primer • Can be enhanced with extension soak at the end of the PCR cycle (e. g. , 15 -45 min @ 60 or 72 o. C) • Can be reduced with new polymerase • Best if there is NOT a mixture of “+/- A” peaks A A • (Clark, J. NAR 16: 9677, Hu. 1993. DNA and Cell Biol. 12: 763. )

Non template directed nucleotide addition to blunt ends • A property of the Taq (and other DNA polymerases), not specific to STRs where an extra nucleotide is added to the 3'OH end of blunt ended double stranded DNA Problem when it is not 100% because peaks (bands) are split (two peaks for the same product, one base pair apart). It is sequence specific, so not all loci will exhibit, and is effected by rxn conditions (eg Mg 2+). • For STRs resolved by adding an extension at the end of thermal cycling. The extension to favor nt+ is currently done at 60 C for 30 minutes. The lower temp is used to reduce 'breathing' between the template and extending strand. The choice of primer sequence can influence the amount of nt+.

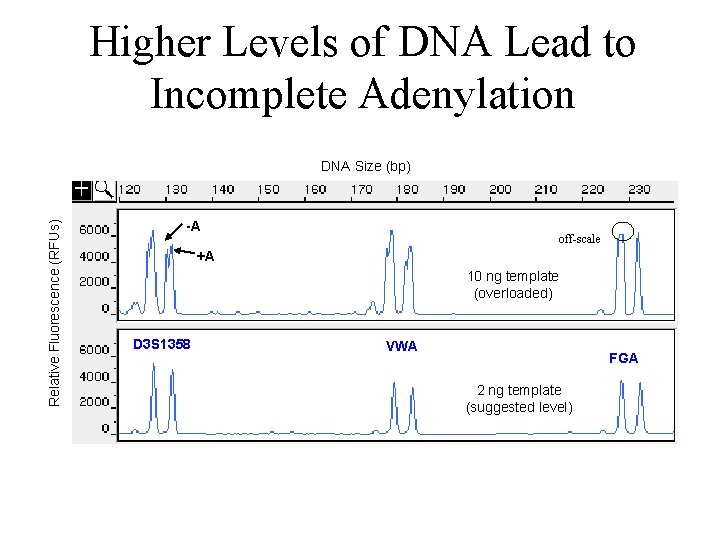
Higher Levels of DNA Lead to Incomplete Adenylation Relative Fluorescence (RFUs) DNA Size (bp) -A off-scale +A 10 ng template (overloaded) D 3 S 1358 VWA FGA 2 ng template (suggested level)

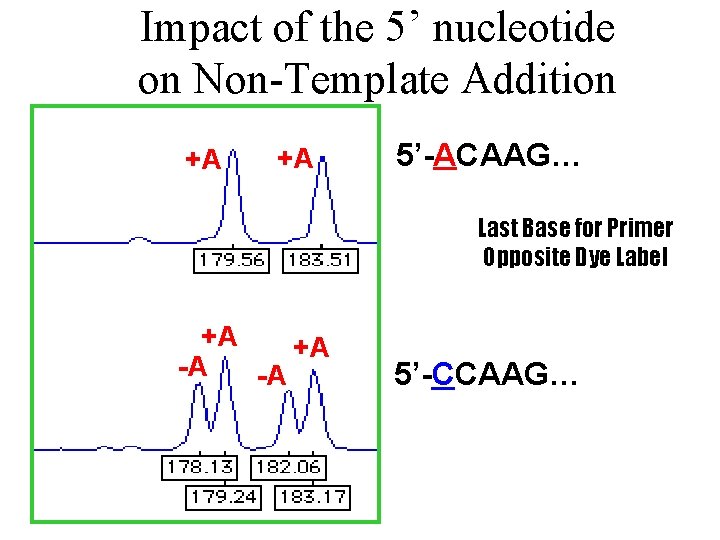
Impact of the 5’ nucleotide on Non-Template Addition +A +A 5’-ACAAG… Last Base for Primer Opposite Dye Label +A +A -A -A 5’-CCAAG…

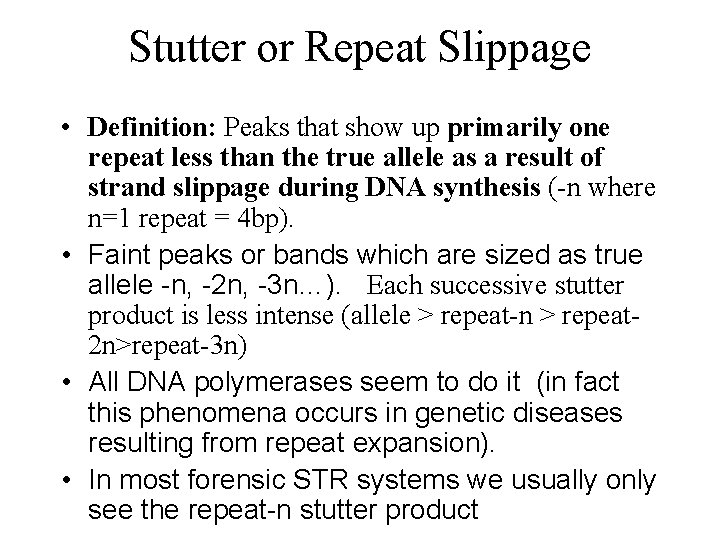
Stutter or Repeat Slippage • Definition: Peaks that show up primarily one repeat less than the true allele as a result of strand slippage during DNA synthesis (-n where n=1 repeat = 4 bp). • Faint peaks or bands which are sized as true allele -n, -2 n, -3 n…). Each successive stutter product is less intense (allele > repeat-n > repeat 2 n>repeat-3 n) • All DNA polymerases seem to do it (in fact this phenomena occurs in genetic diseases resulting from repeat expansion). • In most forensic STR systems we usually only see the repeat-n stutter product

Stutter as it correlates to allele size (eg number of repeats) • Levels of repeat slippage vary for different loci and even for the different alleles of a particular locus. • Amount of repeat slippage appears to be greater in larger alleles with more repeats and less in those that are smaller. Longer repeat regions generate more stutter. That is, a 20 repeat allele will generally have more stutter than a 10 repeat allele • Amount of slippage for a given sized allele

Stutter as it correlates to unit size (eg the number of bases in a single repeat) • Stutter is not as bad with larger repeat unit sizes. • Very bad with small size- di-nucleotide repeats. • Not as bad with larger size - tetra and penta nucleotide repeats • (dinucleotides > tri- > tetra- > penta-)

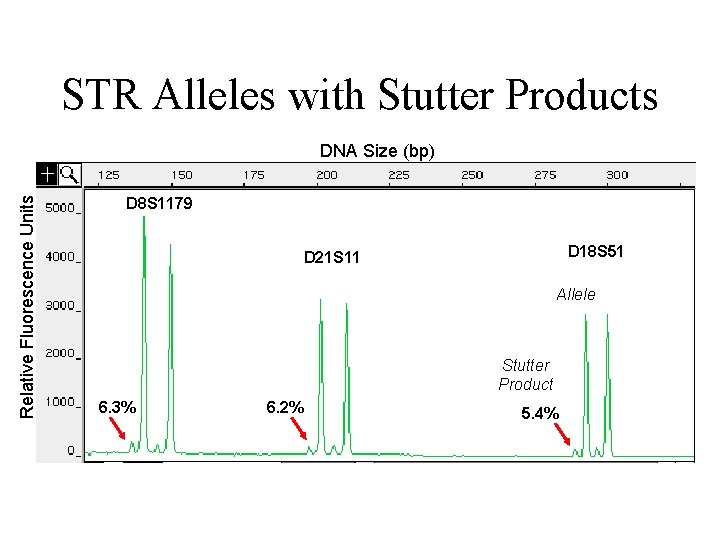
STR Alleles with Stutter Products Relative Fluorescence Units DNA Size (bp) D 8 S 1179 D 18 S 51 D 21 S 11 Allele Stutter Product 6. 3% 6. 2% 5. 4%

Microvariant Alleles • Not alleles have full length repeat units • Alleles with partial repeat units are designated by the number of full repeats and then a decimal point followed by the number of bases in the partial repeat • Example: TH 01 9. 3 allele • (AATG)6(-ATG)(AATG)3

Microvariants • Defined as alleles that are not exact multiples of the basic repeat motif or sequence variants of the repeat motif or both • May exist as insertion, deletion, or base change • Sequence variation can occur within repeat, in the flanking region, or in a primer binding site

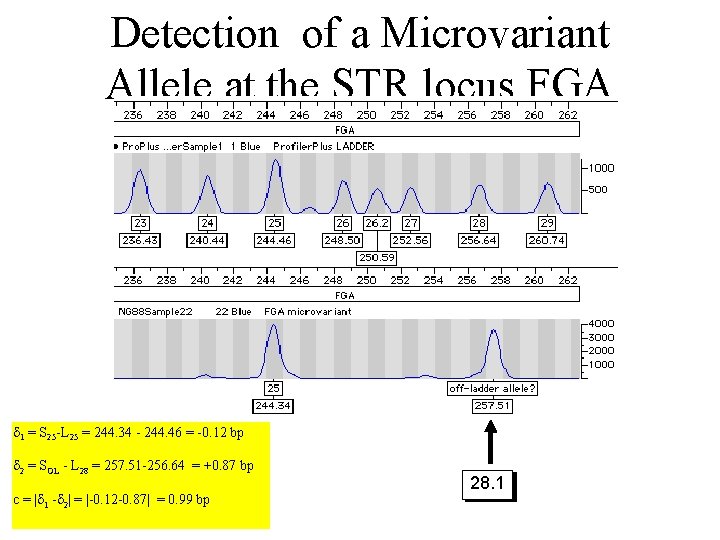
Detection of a Microvariant Allele at the STR locus FGA 1 = S 25 -L 25 = 244. 34 - 244. 46 = -0. 12 bp 2 = SOL - L 28 = 257. 51 -256. 64 = +0. 87 bp c = | 1 - 2| = |-0. 12 -0. 87| = 0. 99 bp 28. 1

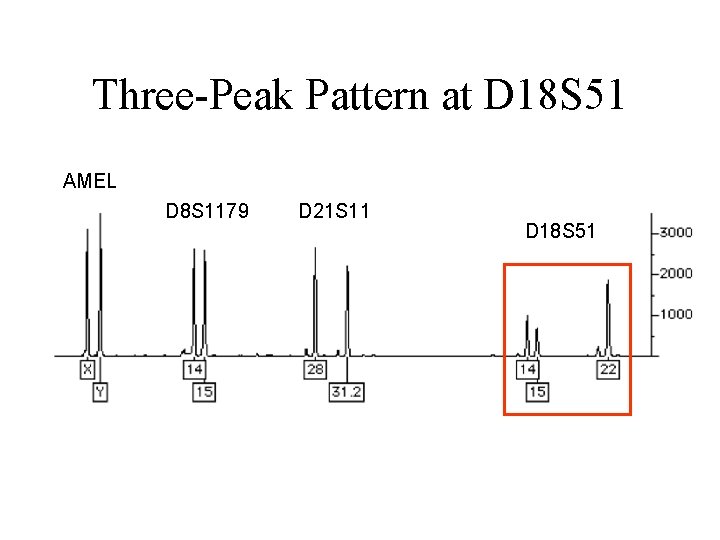
Three-Peak Pattern at D 18 S 51 AMEL D 8 S 1179 D 21 S 11 D 18 S 51

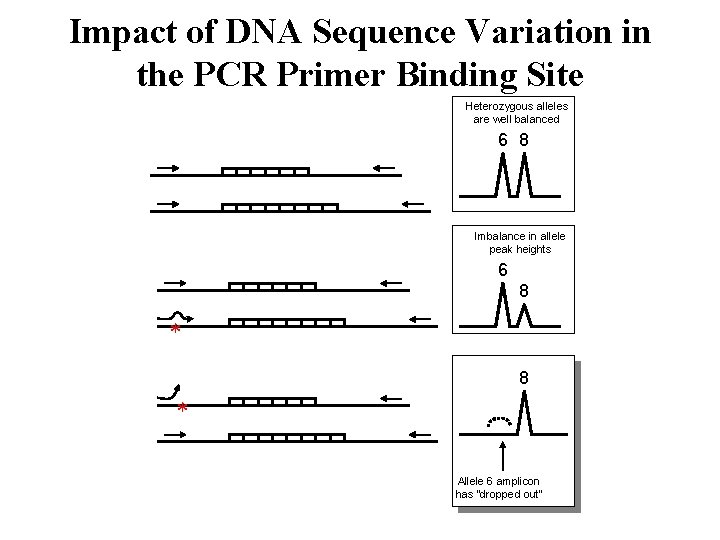
Null Alleles • Allele is present in the DNA sample but fails to be amplified due to a nucleotide change in a primer binding site • Allele dropout is a problem because a heterozygous sample appears falsely as a homozygote • Two PCR primer sets can yield different results on samples originating from the same source • This phenomenon impacts DNA databases • Large concordance studies are typically performed prior to use of new STR kits

Impact of DNA Sequence Variation in the PCR Primer Binding Site Heterozygous alleles are well balanced 6 8 Imbalance in allele peak heights 6 8 * Allele 6 amplicon has “dropped out”

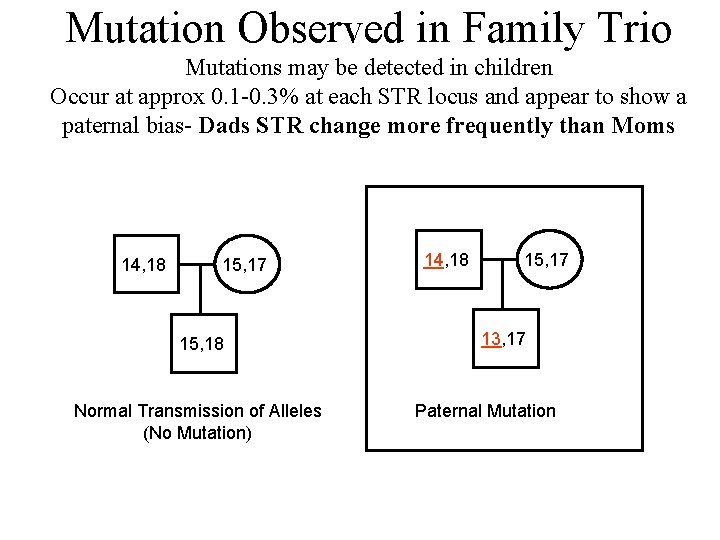
Mutation Observed in Family Trio Mutations may be detected in children Occur at approx 0. 1 -0. 3% at each STR locus and appear to show a paternal bias- Dads STR change more frequently than Moms 14, 18 15, 17 15, 18 Normal Transmission of Alleles (No Mutation) 14, 18 15, 17 13, 17 Paternal Mutation

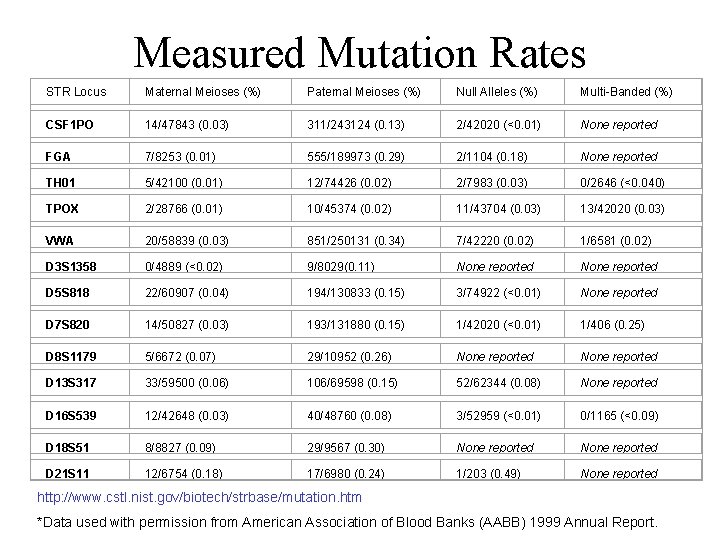
Measured Mutation Rates STR Locus Maternal Meioses (%) Paternal Meioses (%) Null Alleles (%) Multi-Banded (%) CSF 1 PO 14/47843 (0. 03) 311/243124 (0. 13) 2/42020 (<0. 01) None reported FGA 7/8253 (0. 01) 555/189973 (0. 29) 2/1104 (0. 18) None reported TH 01 5/42100 (0. 01) 12/74426 (0. 02) 2/7983 (0. 03) 0/2646 (<0. 040) TPOX 2/28766 (0. 01) 10/45374 (0. 02) 11/43704 (0. 03) 13/42020 (0. 03) VWA 20/58839 (0. 03) 851/250131 (0. 34) 7/42220 (0. 02) 1/6581 (0. 02) D 3 S 1358 0/4889 (<0. 02) 9/8029(0. 11) None reported D 5 S 818 22/60907 (0. 04) 194/130833 (0. 15) 3/74922 (<0. 01) None reported D 7 S 820 14/50827 (0. 03) 193/131880 (0. 15) 1/42020 (<0. 01) 1/406 (0. 25) D 8 S 1179 5/6672 (0. 07) 29/10952 (0. 26) None reported D 13 S 317 33/59500 (0. 06) 106/69598 (0. 15) 52/62344 (0. 08) None reported D 16 S 539 12/42648 (0. 03) 40/48760 (0. 08) 3/52959 (<0. 01) 0/1165 (<0. 09) D 18 S 51 8/8827 (0. 09) 29/9567 (0. 30) None reported D 21 S 11 12/6754 (0. 18) 17/6980 (0. 24) 1/203 (0. 49) None reported http: //www. cstl. nist. gov/biotech/strbase/mutation. htm *Data used with permission from American Association of Blood Banks (AABB) 1999 Annual Report.

Review of STRs Intro to STRs – Head to tail arrangements 4 bp repeat units – Polymorphic, Common, Stably Inherited, Implicated in Diseases – Advantages- Discrete, Small- less prone to PA, Useful on highly degraded DNA, Ability to Multiplex , Provide powerful discrimination. – STR biological artifacts- stutter, adenylation, microvariants, null alleles, mutations – Results are interpreted by reproducibility, size of the resulting fragment, spectral properties, stutter, and size of peak (balance within and among loci). – Multiplexing STR loci provide powerful discrimination