ICCS enewsletter CSI Summer 2011 Jeannine T Holden








- Slides: 23

ICCS e-newsletter CSI Summer 2011 Jeannine T. Holden, MD Emory University School of Medicine Atlanta, GA

Case History The patient is a 35 year old male with a history of B lymphoblastic leukemia/lymphoma diagnosed several years prior to the present study and treated with chemotherapy. Both original diagnosis and treatment were performed at another institution. Patient is reportedly doing well clinically, and the present study is routine monitoring. His peripheral blood counts are all within normal limits. The patient does report a recent two day history of malaise and upper respiratory symptoms that are attributed to viral infection.

Questions… • Is there any evidence of residual/recurrent B lymphoblastic leukemia/lymphoma? • Is normal hematopoietic activity intact? • Is there any evidence of other diseases? • Are there any other problems with this case?

Flow cytometric immunophenotyping • Bone marrow aspirate • Acquired on a FACSCanto cytometer using Diva software, FCS 2. 0 formatted listmode data • Fourteen four-color tubes • Tubes #3 through #14 gated at acquisition (enriching for mononuclear cells) • Files are labeled ICCS 001 through ICCS 014

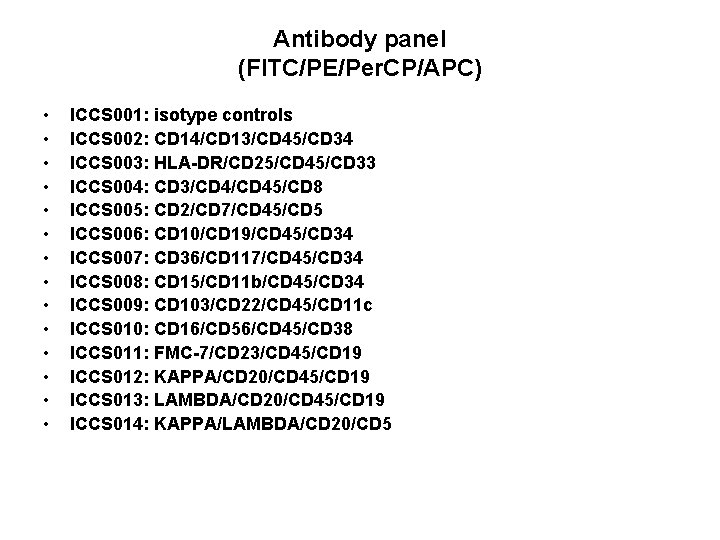
Antibody panel (FITC/PE/Per. CP/APC) • • • • ICCS 001: isotype controls ICCS 002: CD 14/CD 13/CD 45/CD 34 ICCS 003: HLA-DR/CD 25/CD 45/CD 33 ICCS 004: CD 3/CD 45/CD 8 ICCS 005: CD 2/CD 7/CD 45/CD 5 ICCS 006: CD 10/CD 19/CD 45/CD 34 ICCS 007: CD 36/CD 117/CD 45/CD 34 ICCS 008: CD 15/CD 11 b/CD 45/CD 34 ICCS 009: CD 103/CD 22/CD 45/CD 11 c ICCS 010: CD 16/CD 56/CD 45/CD 38 ICCS 011: FMC-7/CD 23/CD 45/CD 19 ICCS 012: KAPPA/CD 20/CD 45/CD 19 ICCS 013: LAMBDA/CD 20/CD 45/CD 19 ICCS 014: KAPPA/LAMBDA/CD 20/CD 5

Antibody panel (FITC/PE/Per. CP/APC) Ignore high background staining with this reagent in • this ICCS 001: case! isotype controls • • • • ICCS 002: CD 14/CD 13/CD 45/CD 34 ICCS 003: HLA-DR/CD 25/CD 45/CD 33 ICCS 004: CD 3/CD 45/CD 8 ICCS 005: CD 2/CD 7/CD 45/CD 5 ICCS 006: CD 10/CD 19/CD 45/CD 34 ICCS 007: CD 36/CD 117/CD 45/CD 34 ICCS 008: CD 15/CD 11 B/CD 45/CD 34 ICCS 009: CD 103/CD 22/CD 45/CD 11 c ICCS 010: CD 16/CD 56/CD 45/CD 38 ICCS 011: FMC-7/CD 23/CD 45/CD 19 ICCS 012: KAPPA/CD 20/CD 45/CD 19 ICCS 013: LAMBDA/CD 20/CD 45/CD 19 ICCS 014: KAPPA/LAMBDA/CD 20/CD 5

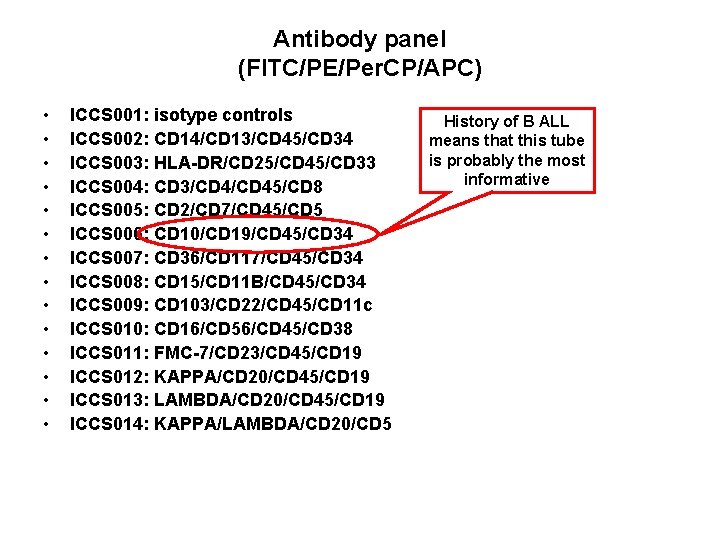
Antibody panel (FITC/PE/Per. CP/APC) • • • • ICCS 001: isotype controls ICCS 002: CD 14/CD 13/CD 45/CD 34 ICCS 003: HLA-DR/CD 25/CD 45/CD 33 ICCS 004: CD 3/CD 45/CD 8 ICCS 005: CD 2/CD 7/CD 45/CD 5 ICCS 006: CD 10/CD 19/CD 45/CD 34 ICCS 007: CD 36/CD 117/CD 45/CD 34 ICCS 008: CD 15/CD 11 B/CD 45/CD 34 ICCS 009: CD 103/CD 22/CD 45/CD 11 c ICCS 010: CD 16/CD 56/CD 45/CD 38 ICCS 011: FMC-7/CD 23/CD 45/CD 19 ICCS 012: KAPPA/CD 20/CD 45/CD 19 ICCS 013: LAMBDA/CD 20/CD 45/CD 19 ICCS 014: KAPPA/LAMBDA/CD 20/CD 5 History of B ALL means that this tube is probably the most informative

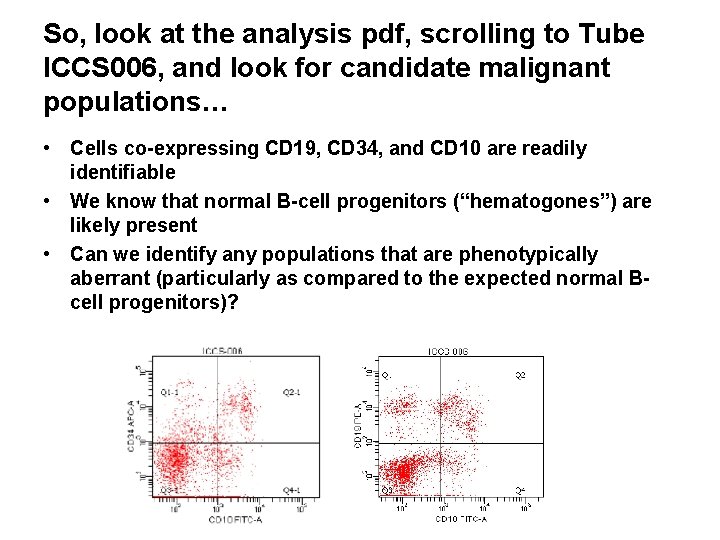
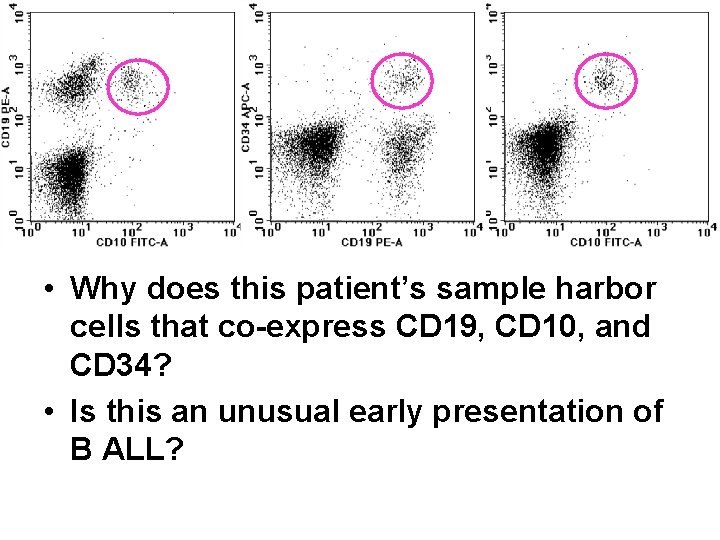
So, look at the analysis pdf, scrolling to Tube ICCS 006, and look for candidate malignant populations… • Cells co-expressing CD 19, CD 34, and CD 10 are readily identifiable • We know that normal B-cell progenitors (“hematogones”) are likely present • Can we identify any populations that are phenotypically aberrant (particularly as compared to the expected normal Bcell progenitors)?

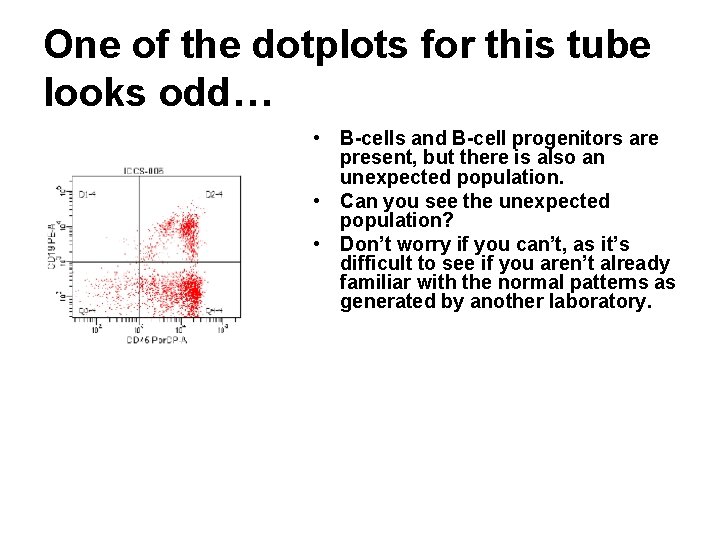
One of the dotplots for this tube looks odd… • B-cells and B-cell progenitors are present, but there is also an unexpected population. • Can you see the unexpected population? • Don’t worry if you can’t, as it’s difficult to see if you aren’t already familiar with the normal patterns as generated by another laboratory.

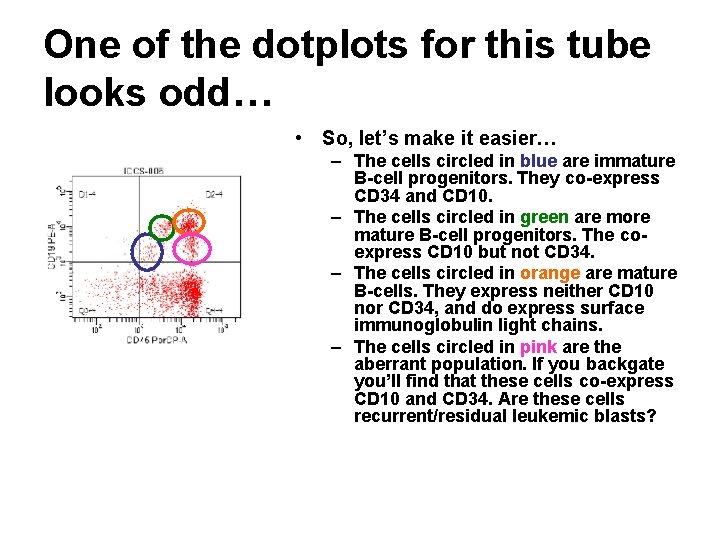
One of the dotplots for this tube looks odd… • So, let’s make it easier… – The cells circled in blue are immature B-cell progenitors. They co-express CD 34 and CD 10. – The cells circled in green are more mature B-cell progenitors. The coexpress CD 10 but not CD 34. – The cells circled in orange are mature B-cells. They express neither CD 10 nor CD 34, and do express surface immunoglobulin light chains. – The cells circled in pink are the aberrant population. If you backgate you’ll find that these cells co-express CD 10 and CD 34. Are these cells recurrent/residual leukemic blasts?

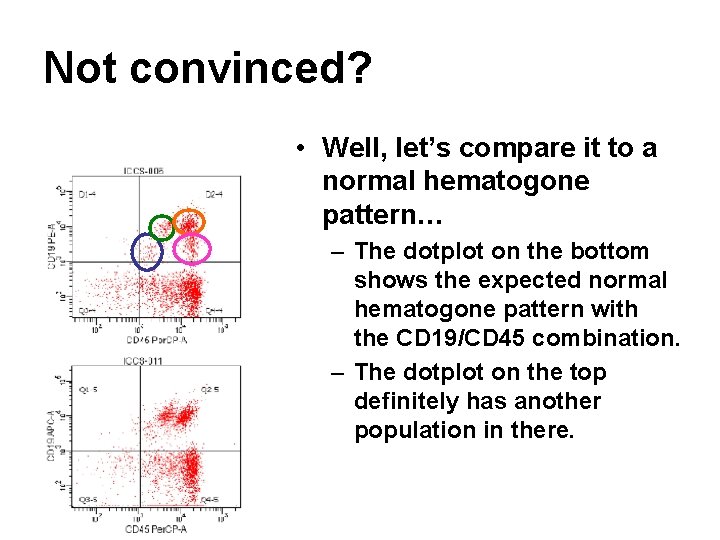
Not convinced? • Well, let’s compare it to a normal hematogone pattern… – The dotplot on the bottom shows the expected normal hematogone pattern with the CD 19/CD 45 combination. – The dotplot on the top definitely has another population in there.

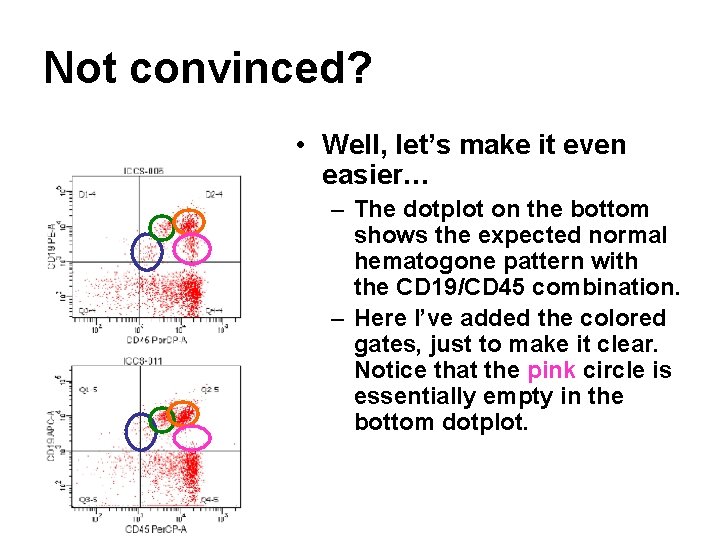
Not convinced? • Well, let’s make it even easier… – The dotplot on the bottom shows the expected normal hematogone pattern with the CD 19/CD 45 combination. – Here I’ve added the colored gates, just to make it clear. Notice that the pink circle is essentially empty in the bottom dotplot.

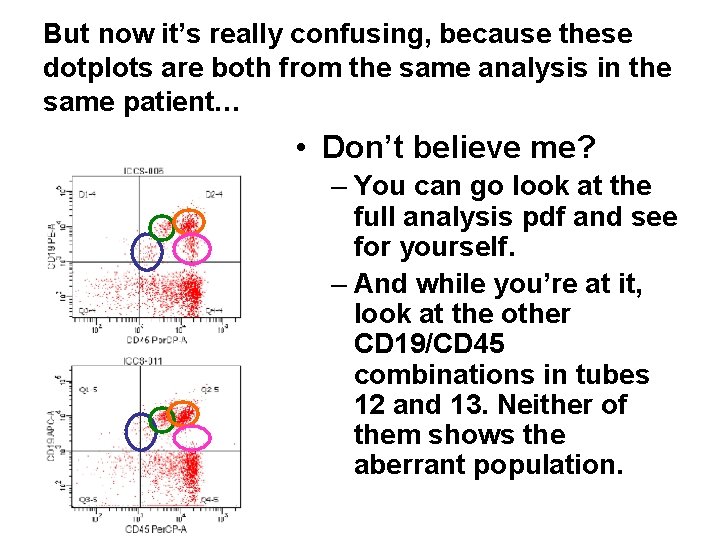
But now it’s really confusing, because these dotplots are both from the same analysis in the same patient… • Don’t believe me? – You can go look at the full analysis pdf and see for yourself. – And while you’re at it, look at the other CD 19/CD 45 combinations in tubes 12 and 13. Neither of them shows the aberrant population.

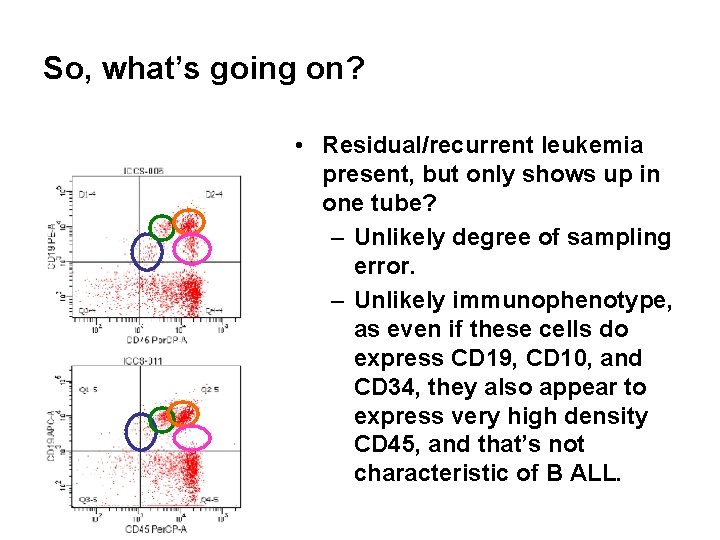
So, what’s going on? • Residual/recurrent leukemia present, but only shows up in one tube? – Unlikely degree of sampling error. – Unlikely immunophenotype, as even if these cells do express CD 19, CD 10, and CD 34, they also appear to express very high density CD 45, and that’s not characteristic of B ALL.

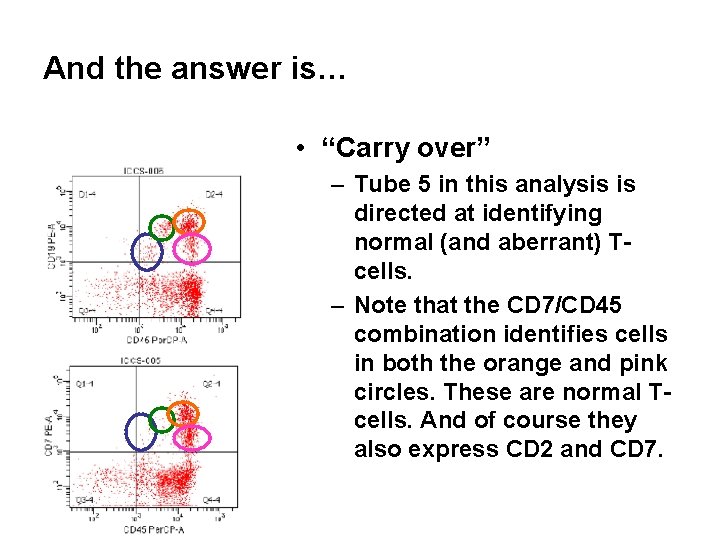
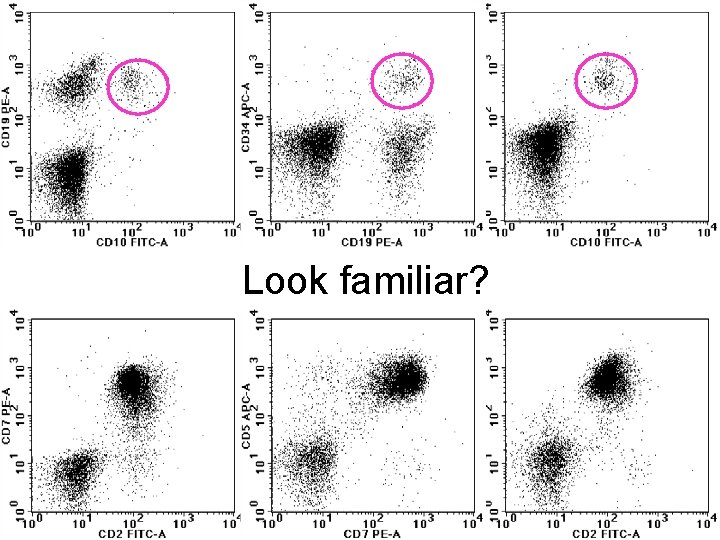
And the answer is… • “Carry over” – Tube 5 in this analysis is directed at identifying normal (and aberrant) Tcells. – Note that the CD 7/CD 45 combination identifies cells in both the orange and pink circles. These are normal Tcells. And of course they also express CD 2 and CD 7.

“Carry over” • Already stained cells from one tube are introduced to another tube of stained cells • Results in overlapping dotplots • Typically identified in sequential tubes • May occur as a result of… – – Pipetting error (unusual) Splashing of one sample into an another Inefficient washing/rinsing of sipper Machine tubing: cells from first tube remain in tubing, acquired with next tube

How do I know for sure that this is “carry over”? • Because I deliberately contaminated tube 6 with cells from tube 5, that’s why. • But here’s another example to show you what it looks like…

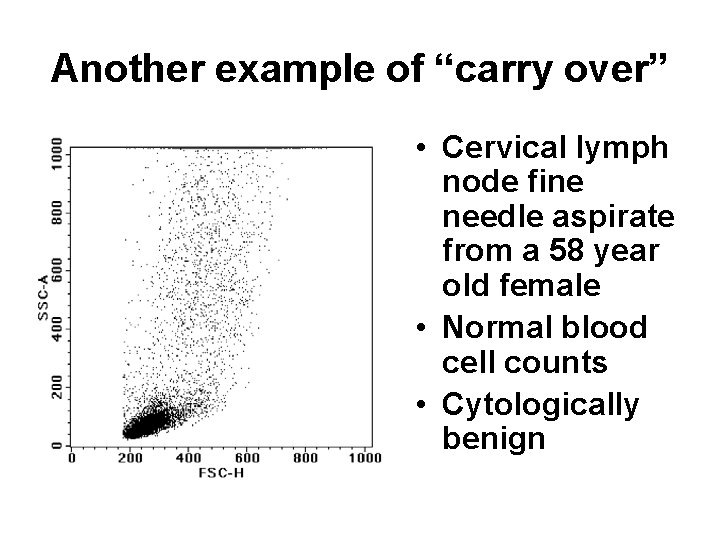
Another example of “carry over” • Cervical lymph node fine needle aspirate from a 58 year old female • Normal blood cell counts • Cytologically benign

• Why does this patient’s sample harbor cells that co-express CD 19, CD 10, and CD 34? • Is this an unusual early presentation of B ALL?

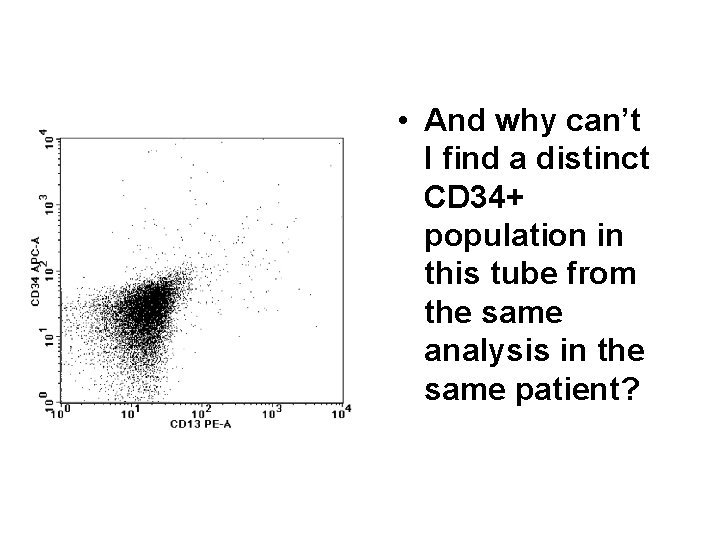
• And why can’t I find a distinct CD 34+ population in this tube from the same analysis in the same patient?

Look familiar?


Conclusions • Exploit panel redundancy to investigate unexpected findings. • Assume that errors will occur. • Consider all information about a case, including the clinical history. • Don’t have the clinical history? Get it.
 La priere du soir de jeannine savard
La priere du soir de jeannine savard Jeannine balfour
Jeannine balfour Jeannine balfour
Jeannine balfour Jeannine balfour
Jeannine balfour Cbc normal values
Cbc normal values Iccs
Iccs Iata iccs
Iata iccs Iccs 2016 cytometry
Iccs 2016 cytometry Iccs denver
Iccs denver Who is sunny in catcher in the rye
Who is sunny in catcher in the rye Examples of holden caulfield being an unreliable narrator
Examples of holden caulfield being an unreliable narrator Trigon
Trigon Catcher in the rye sunny
Catcher in the rye sunny Janice miner holden
Janice miner holden What does holden fantasize about
What does holden fantasize about Dhiraj holden
Dhiraj holden Georgina holden
Georgina holden The catcher in the rye timeline review
The catcher in the rye timeline review Robert burns catcher in the rye
Robert burns catcher in the rye Why didn't holden throw the snowball
Why didn't holden throw the snowball Connie holden
Connie holden Why does holden attack stradlater?
Why does holden attack stradlater? Joanna holden
Joanna holden Chapter 26 catcher in the rye
Chapter 26 catcher in the rye










































