Hemoproteins Axial Ligands and Functions From Ccile Claude




- Slides: 9

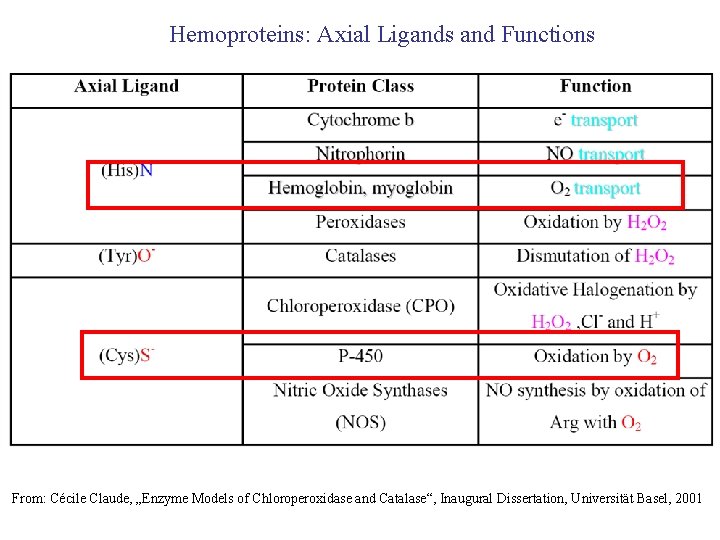
Hemoproteins: Axial Ligands and Functions From: Cécile Claude, „Enzyme Models of Chloroperoxidase and Catalase“, Inaugural Dissertation, Universität Basel, 2001

F 8390 Metalloproteins: Structure and Function 1. Introduction 1. 1. Metalloproteins: Functions in Biological Chemistry 1. 2. Some fundamental metal sites in metalloproteins 2. Mononuclear zinc enzymes: Carbonic anhydrase 3. Metalloproteins reacting with oxygen 3. 1. Why do aerobic organisms need metalloproteins? 3. 2. Oxygen transport proteins & Oxygenases 3. 2. 1. Hemoglobin, Myoglobin Cytochrome P 450 3. 2. 2. Hemerythrin & Ribonucleotide Reductase R 2 & Methane monooxygenase diiron subunits 3. 2. 3. Hemocyanin & Tyrosinase 4. Electron transfer proteins 4. 1. Iron-sulfur proteins 4. 2. Blue copper proteins 5. Conclusion

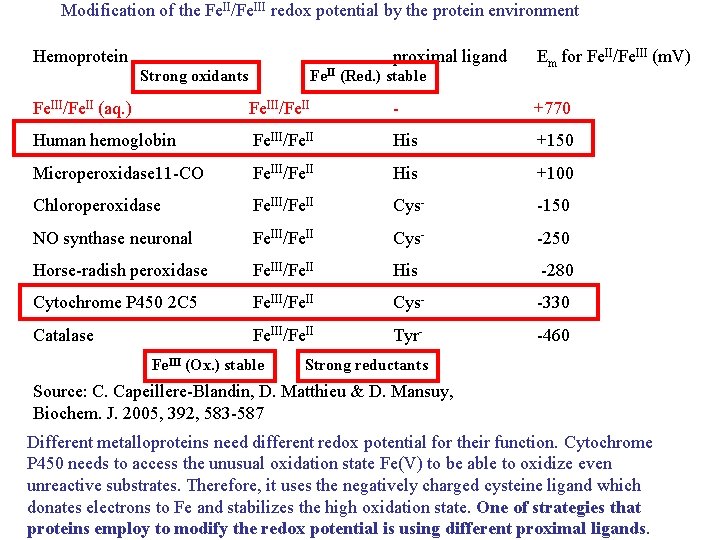
Modification of the Fe. II/Fe. III redox potential by the protein environment Hemoprotein proximal ligand Strong oxidants Fe. II (Red. ) stable Em for Fe. II/Fe. III (m. V) Fe. III/Fe. II (aq. ) Fe. III/Fe. II - +770 Human hemoglobin Fe. III/Fe. II His +150 Microperoxidase 11 -CO Fe. III/Fe. II His +100 Chloroperoxidase Fe. III/Fe. II Cys- -150 NO synthase neuronal Fe. III/Fe. II Cys- -250 Horse-radish peroxidase Fe. III/Fe. II His -280 Cytochrome P 450 2 C 5 Fe. III/Fe. II Cys- -330 Catalase Fe. III/Fe. II Tyr- -460 Fe. III (Ox. ) stable Strong reductants Source: C. Capeillere-Blandin, D. Matthieu & D. Mansuy, Biochem. J. 2005, 392, 583 -587 Different metalloproteins need different redox potential for their function. Cytochrome P 450 needs to access the unusual oxidation state Fe(V) to be able to oxidize even unreactive substrates. Therefore, it uses the negatively charged cysteine ligand which donates electrons to Fe and stabilizes the high oxidation state. One of strategies that proteins employ to modify the redox potential is using different proximal ligands.

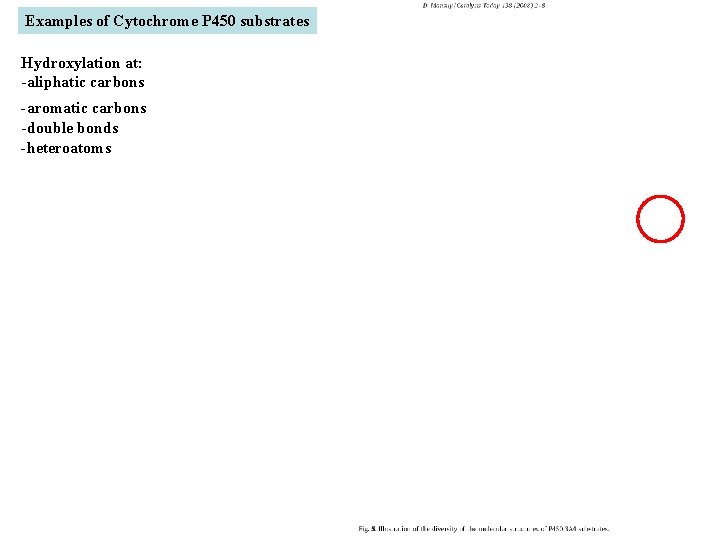
Examples of Cytochrome P 450 substrates Hydroxylation at: -aliphatic carbons -aromatic carbons -double bonds -heteroatoms local anesthetic steroid hormone carcinogen from fungi antibiotic Alkaloid from Taxus brevifolia, potent anti-cancer drug

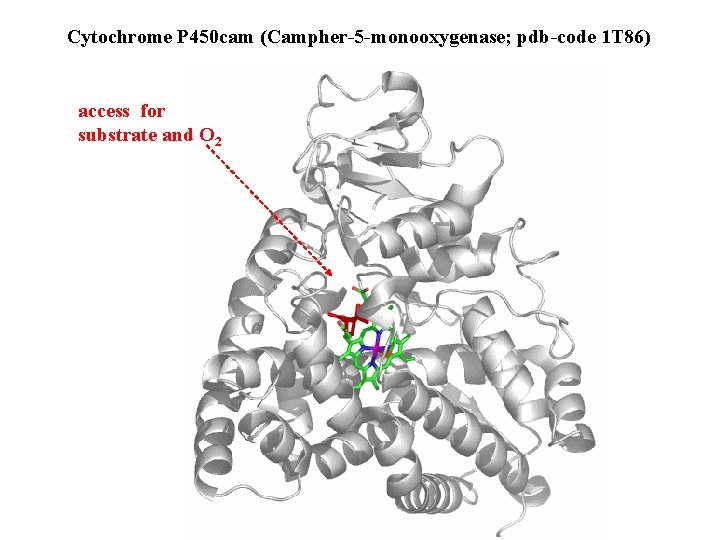
Cytochrome P 450 cam (Campher-5 -monooxygenase; pdb-code 1 T 86) access for substrate and O 2


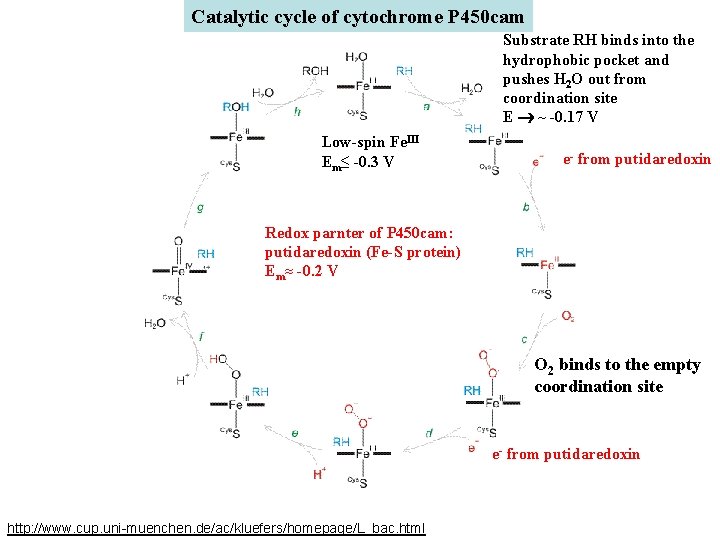
Catalytic cycle of cytochrome P 450 cam Substrate RH binds into the hydrophobic pocket and pushes H 2 O out from coordination site E ~ -0. 17 V Low-spin Fe. III Em≤ -0. 3 V e- from putidaredoxin Redox parnter of P 450 cam: putidaredoxin (Fe-S protein) Em≈ -0. 2 V O 2 binds to the empty coordination site e- from putidaredoxin http: //www. cup. uni-muenchen. de/ac/kluefers/homepage/L_bac. html

Conclusion In many cases, metalloproteins use the same or similar active site for different purposes. The strategies to confer a particular activity to a given site include - Allowing/disallowing access of substrates to the active site (including the dynamics of diffusion of substrate/product) -Modifying the electrostatic potential by mutating the amino acids coordinated to the metal or surrounding the binding pocket

Practical training - Download from the pdb database the structures of bacterial cytochrome P 450 cam 1 t 86 and 1 dz 8 http: //www. rcsb. org/pdb/home. do - Display the structures using VMD - Use the command „chain A“ in Graphics/Representation“ to display only the monomer A - Use the command „chain A and resname HEM“ in Graphics/Representation“ to highlight the heme group - Observe whether the two crystal structures contain the campher and/or oxygen molecule trapped near the active site - Use the command „chain A and resname CAM“ in Graphics/Representation“ to highlight the campher molecule -Use the command „chain A and resname OXY“ in Graphics/Representation“ to highlight the O 2 molecule - Examine how the two carboxylate groups of heme are anchored in the protein backbone - Examine how the campher substrate is fixed in the acess channel