GOA Looking after GO annotations Emily Dimmer Gene










- Slides: 39

GOA: Looking after GO annotations Emily Dimmer Gene Ontology Annotation (GOA) Database European Bioinformatics Institute Cambridge UK EBI is an Outstation of the European Molecular Biology Laboratory.

E. Coli hub http: //www. geneontology. org Reactome 2 EMBRACE Workshop 7 -9 th November 2007

Gene Ontology Annotation (GOA) Database • Member of the GO Consortium since 2001 • Largest open-source contributor of annotations to GO • Provides annotation for more than 139, 000 species • GOA’s priority is to annotate the human proteome • GOA is responsible for human, chicken and bovine annotations in the GO Consortium 3 EMBRACE Workshop 7 -9 th November 2007

GOA Group GOA office EMBL-EBI Wellcome Trust Genome Campus, Hinxton, Cambridge, UK 4 goa@ebi. ac. uk EMBRACE Workshop 7 -9 th November 2007

GOA Group Emily Dimmer Evelyn Camon (GOA coordinator) (senior GOA curator) Rachael Huntley (GOA curator) Daniel Barrell (GOA file releases & database) David Binns (Quick. GO, protein 2 go tools) Along with the help of Uni. Prot curators at the EBI, Uni. Prot controlled vocabularies, HAMAP group, Inter. Pro group, Int. Act curators, the IPI group, Ensembl, other EBI groups …and of course the GO editors and the other GO Consortium annotation groups 5 EMBRACE Workshop 7 -9 th November 2007

How does GOA annotate to the GO ? Electronic Annotation Manual Annotation • Both these methods have their advantages • They can be easily distinguished by the evidence code used. 6 EMBRACE Workshop 7 -9 th November 2007

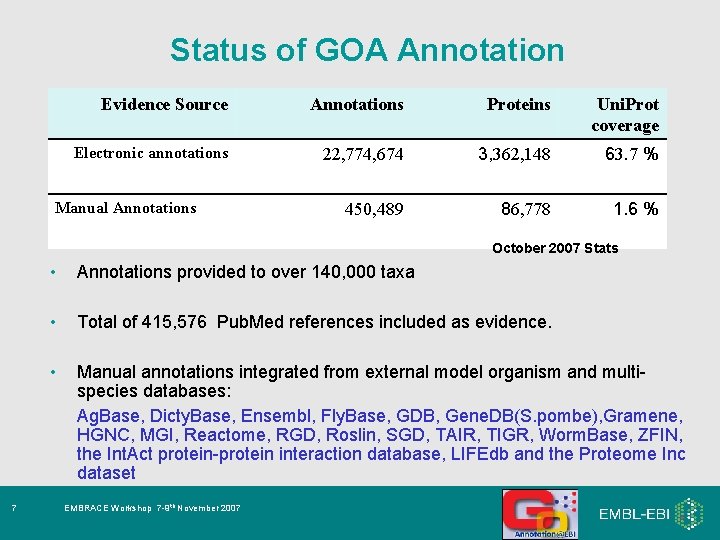
Status of GOA Annotation Evidence Source Annotations Proteins Uni. Prot coverage Electronic annotations 22, 774, 674 3, 362, 148 63. 7 % 450, 489 86, 778 1. 6 % Manual Annotations October 2007 Stats 7 • Annotations provided to over 140, 000 taxa • Total of 415, 576 Pub. Med references included as evidence. • Manual annotations integrated from external model organism and multispecies databases: Ag. Base, Dicty. Base, Ensembl, Fly. Base, GDB, Gene. DB(S. pombe), Gramene, HGNC, MGI, Reactome, RGD, Roslin, SGD, TAIR, TIGR, Worm. Base, ZFIN, the Int. Act protein-protein interaction database, LIFEdb and the Proteome Inc dataset EMBRACE Workshop 7 -9 th November 2007

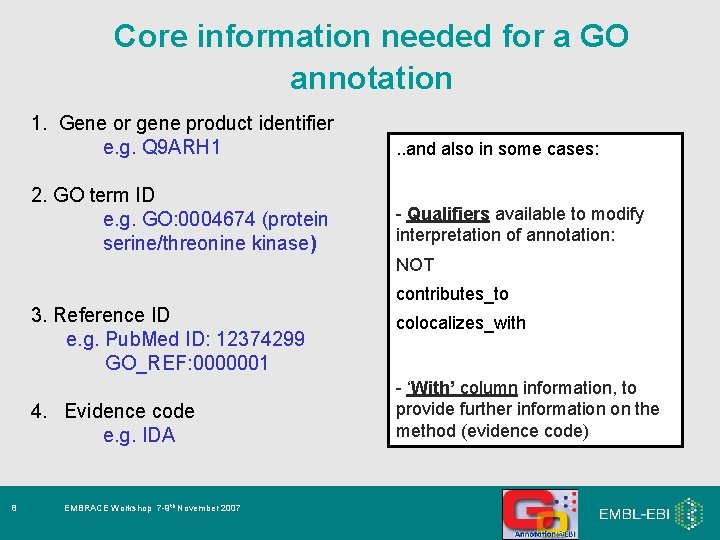
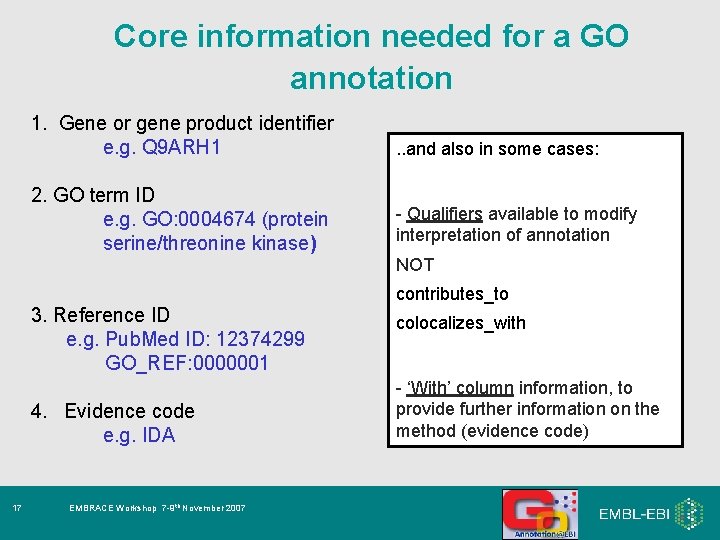
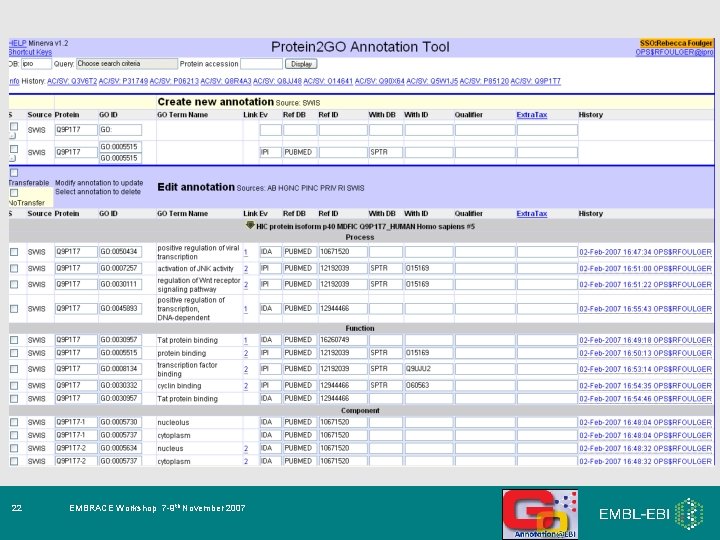
Core information needed for a GO annotation 1. Gene or gene product identifier e. g. Q 9 ARH 1 . . and also in some cases: 2. GO term ID e. g. GO: 0004674 (protein serine/threonine kinase) - Qualifiers available to modify interpretation of annotation: NOT contributes_to 3. Reference ID e. g. Pub. Med ID: 12374299 GO_REF: 0000001 4. Evidence code e. g. IDA 8 EMBRACE Workshop 7 -9 th November 2007 colocalizes_with - ‘With’ column information, to provide further information on the method (evidence code)

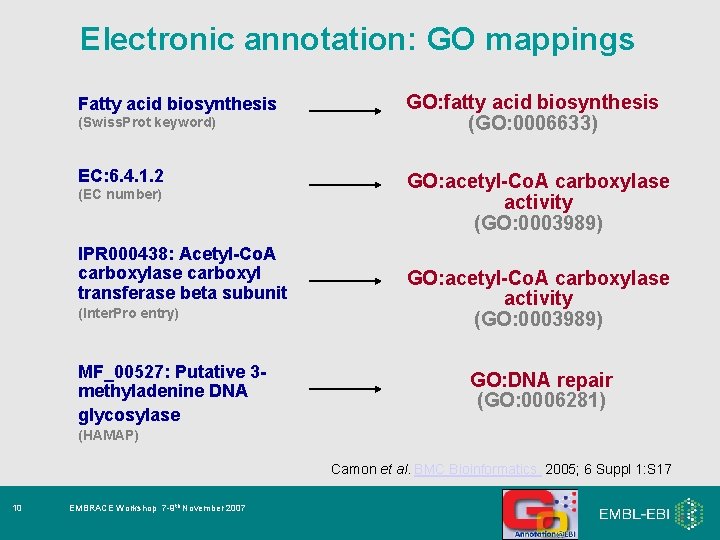
Electronic Annotation • A number of different techniques used by different GO Consortium annotation groups. • All resulting annotations must be high-quality and provide an explanation of the method (GO_REF) 1. Mapping of external concepts to GO terms 2. Automatic transfer of annotations to orthologs 9 EMBRACE Workshop 7 -9 th November 2007

Electronic annotation: GO mappings Fatty acid biosynthesis (Swiss. Prot keyword) EC: 6. 4. 1. 2 (EC number) IPR 000438: Acetyl-Co. A carboxylase carboxyl transferase beta subunit (Inter. Pro entry) MF_00527: Putative 3 methyladenine DNA glycosylase GO: fatty acid biosynthesis (GO: 0006633) GO: acetyl-Co. A carboxylase activity (GO: 0003989) GO: DNA repair (GO: 0006281) (HAMAP) Camon et al. BMC Bioinformatics. 2005; 6 Suppl 1: S 17 10 EMBRACE Workshop 7 -9 th November 2007

11 EMBRACE Workshop 7 -9 th November 2007

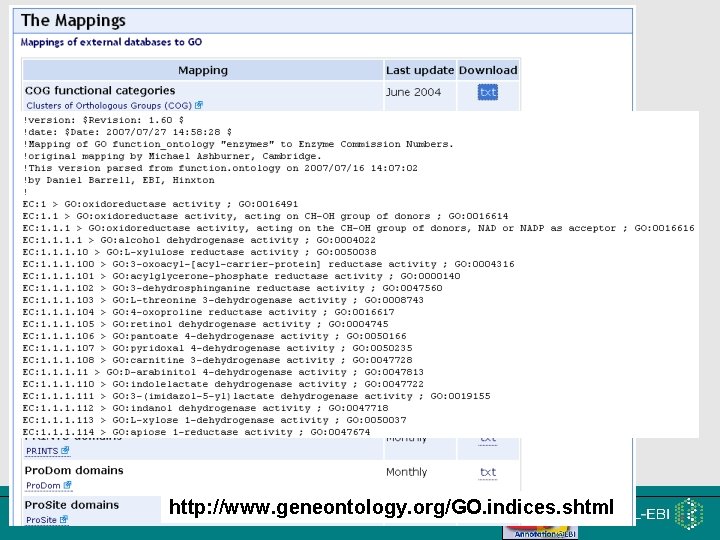
12 http: //www. geneontology. org/GO. indices. shtml EMBRACE Workshop 7 -9 th November 2007

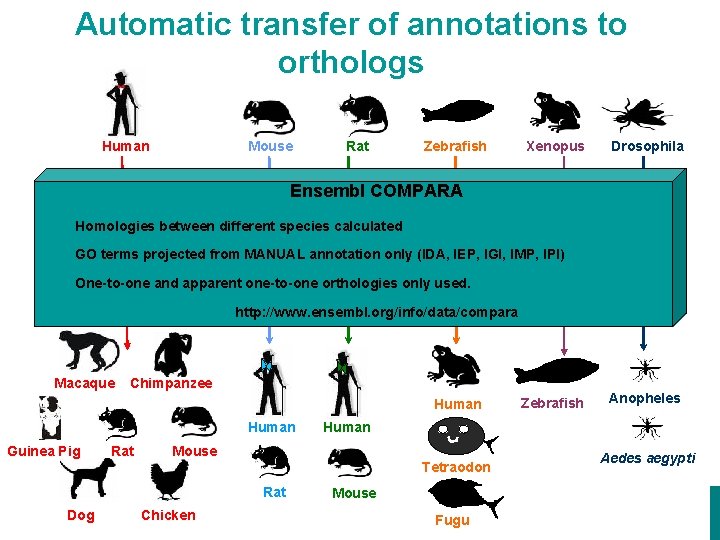
Automatic transfer of annotations to orthologs Human Mouse Rat Zebrafish Xenopus Drosophila Ensembl COMPARA Homologies between different species calculated GO terms projected from MANUAL annotation only (IDA, IEP, IGI, IMP, IPI) One-to-one and apparent one-to-one orthologies only used. http: //www. ensembl. org/info/data/compara Macaque Chimpanzee Human Guinea Pig Rat Mouse EMBRACE Workshop 7 -9 th November 2007 Dog Chicken Anopheles Human Tetraodon 13 Zebrafish Fugu Aedes aegypti

Manual Annotation • High–quality, specific annotations made using: • Peer-reviewed papers • A range of evidence codes to categorize the types of evidence found in a paper • Very time consuming and requires trained biologists 14 EMBRACE Workshop 7 -9 th November 2007

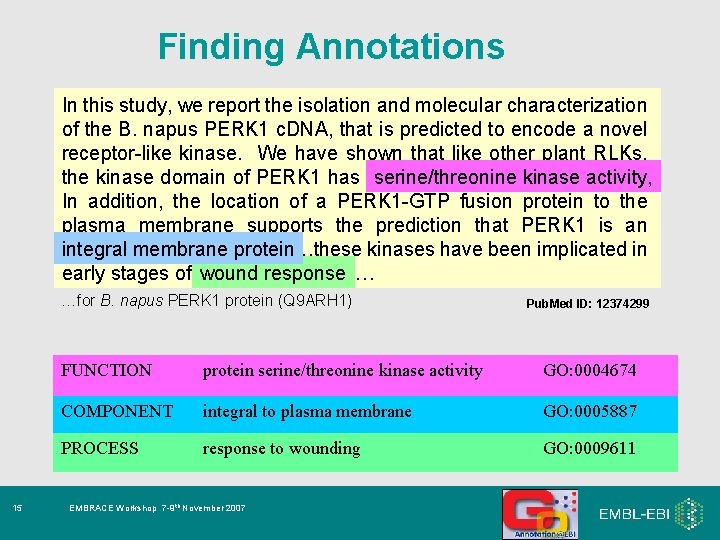
Finding Annotations In this study, we report the isolation and molecular characterization of the B. napus PERK 1 c. DNA, that is predicted to encode a novel receptor-like kinase. We have shown that like other plant RLKs, serine/threoninekinaseactivity, the kinase domain of PERK 1 has serine/threonine In addition, the location of a PERK 1 -GTP fusion protein to the plasma membrane supports the prediction that PERK 1 is an kinases have been implicated in integral membrane protein…these protein early stages of woundresponse… …for B. napus PERK 1 protein (Q 9 ARH 1) 15 Pub. Med ID: 12374299 FUNCTION protein serine/threonine kinase activity GO: 0004674 COMPONENT integral to plasma membrane GO: 0005887 PROCESS response to wounding GO: 0009611 EMBRACE Workshop 7 -9 th November 2007

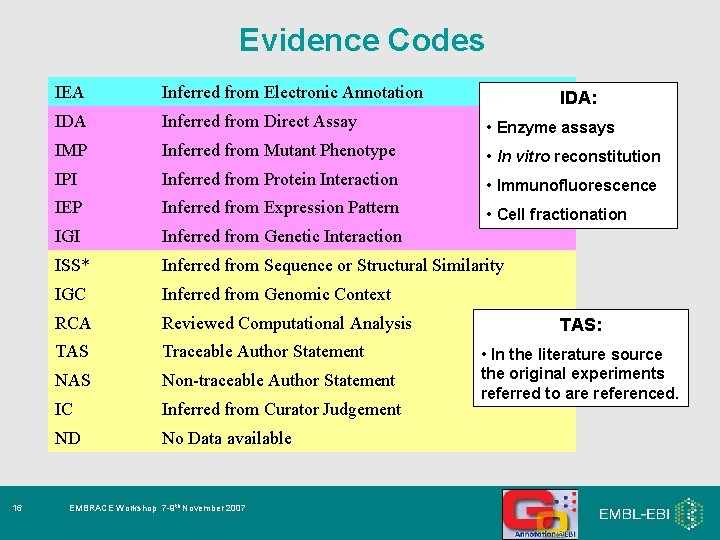
Evidence Codes 16 IEA Inferred from Electronic Annotation IDA Inferred from Direct Assay • Enzyme assays IMP Inferred from Mutant Phenotype • In vitro reconstitution IPI Inferred from Protein Interaction • Immunofluorescence IEP Inferred from Expression Pattern • Cell fractionation IGI Inferred from Genetic Interaction ISS* Inferred from Sequence or Structural Similarity IGC Inferred from Genomic Context RCA Reviewed Computational Analysis TAS Traceable Author Statement NAS Non-traceable Author Statement IC Inferred from Curator Judgement ND No Data available EMBRACE Workshop 7 -9 th November 2007 IDA: TAS: • In the literature source the original experiments referred to are referenced.

Core information needed for a GO annotation 1. Gene or gene product identifier e. g. Q 9 ARH 1 . . and also in some cases: 2. GO term ID e. g. GO: 0004674 (protein serine/threonine kinase) - Qualifiers available to modify interpretation of annotation NOT contributes_to 3. Reference ID e. g. Pub. Med ID: 12374299 GO_REF: 0000001 4. Evidence code e. g. IDA 17 EMBRACE Workshop 7 -9 th November 2007 colocalizes_with - ‘With’ column information, to provide further information on the method (evidence code)

The ‘Qualifier’ Column The Qualifier column is used to modify the interpretation of an annotation. Allowable values are: NOT colocalizes_with contributes_to 18 EMBRACE Workshop 7 -9 th November 2007

The ‘NOT’ qualifier • 'NOT' is used to make an explicit note that the gene product is not associated with the GO term. … particularly important when associating a GO term with a gene product should be avoided (but might otherwise be made, especially by an automated method). e. g. This protein does not have ‘kinase activity’ because it has been found that this protein has a disrupted/missing an ‘ATP binding’ domain. Also used to document conflicting claims in the literature. NOT can be used with ALL three GO Ontologies. 19 EMBRACE Workshop 7 -9 th November 2007

The ‘colocalizes_with’ qualifier • Gene products that are transiently or peripherally associated with an organelle or complex may be annotated to the relevant cellular component term, using the 'colocalizes_with' qualifier. Only used with GO Component Ontology 20 EMBRACE Workshop 7 -9 th November 2007

The ‘contributes_to’ qualifier Where an individual gene product that is part of a complex can be annotated to terms that describe the action (function or process) of the whole complex. i. e. annotating 'to the potential of the complex‘ • distinguishes an individual subunit from complex functions All gene products annotated using 'contributes_to' must also be annotated to a cellular component term representing the complex that possesses the activity. Only used with GO Function Ontology 21 EMBRACE Workshop 7 -9 th November 2007

22 EMBRACE Workshop 7 -9 th November 2007

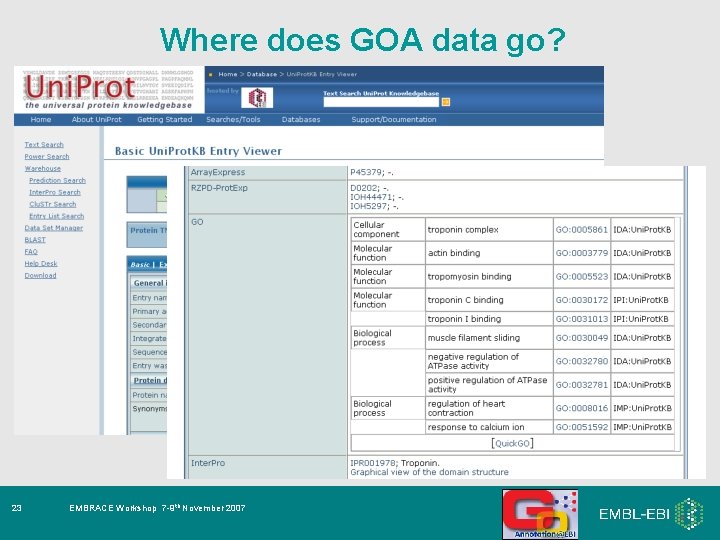
Where does GOA data go? 23 EMBRACE Workshop 7 -9 th November 2007

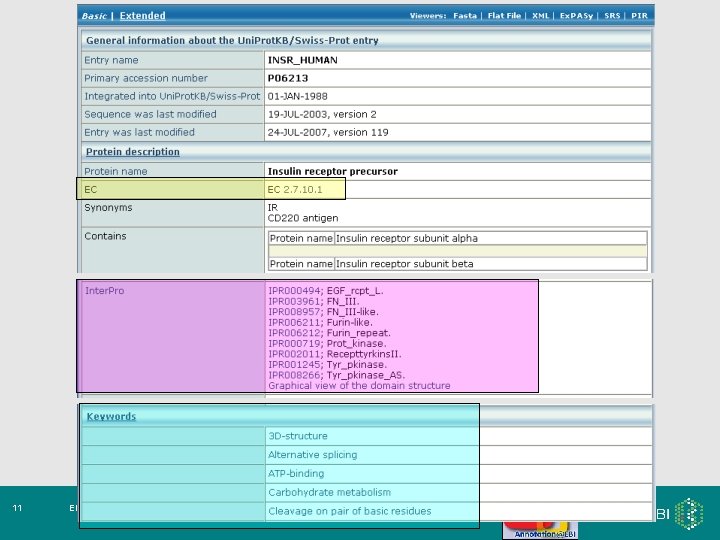
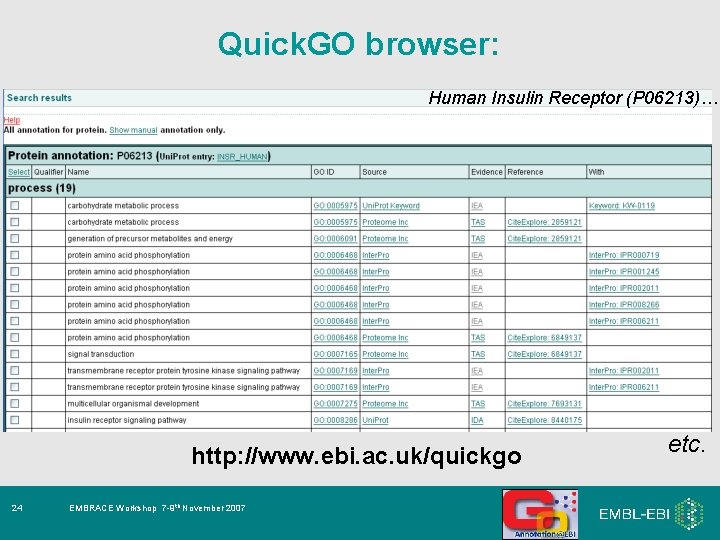
Quick. GO browser: Human Insulin Receptor (P 06213)… http: //www. ebi. ac. uk/quickgo 24 EMBRACE Workshop 7 -9 th November 2007 etc.

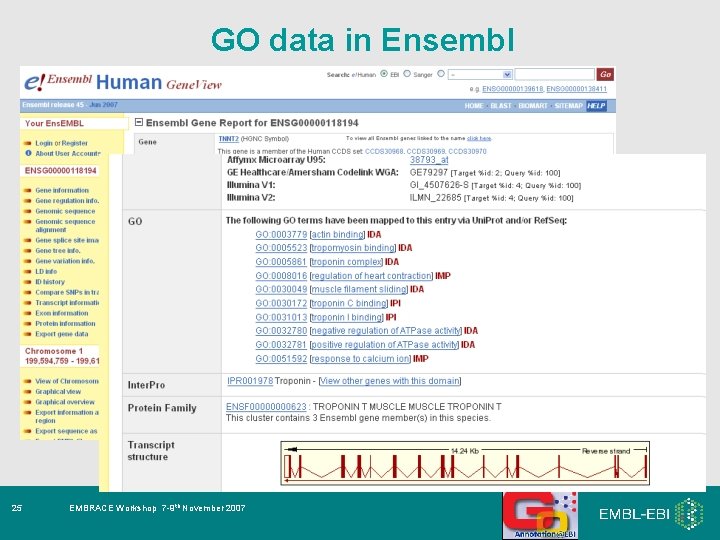
GO data in Ensembl 25 EMBRACE Workshop 7 -9 th November 2007

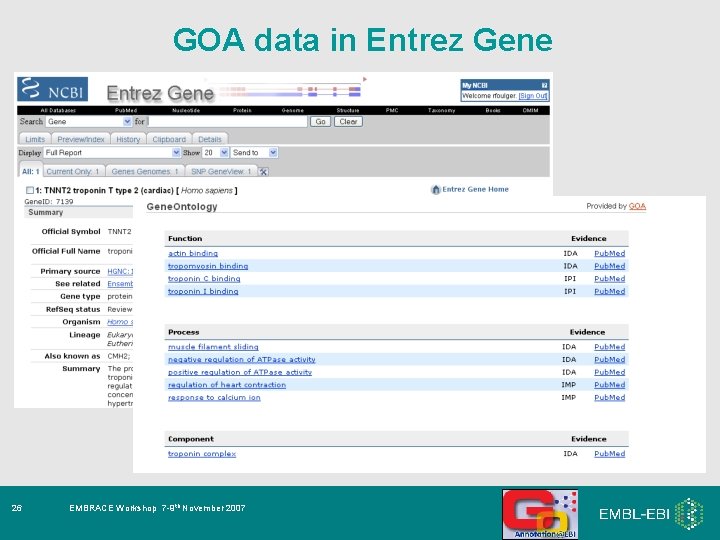
GOA data in Entrez Gene 26 EMBRACE Workshop 7 -9 th November 2007

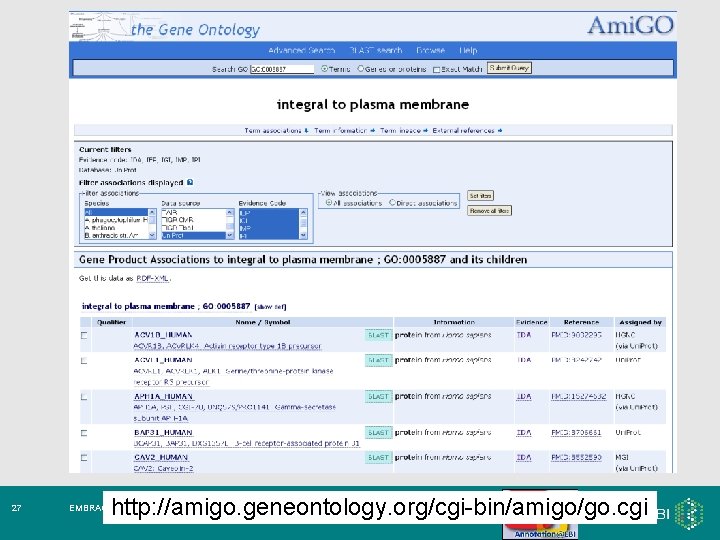
27 http: //amigo. geneontology. org/cgi-bin/amigo/go. cgi EMBRACE Workshop 7 -9 th November 2007

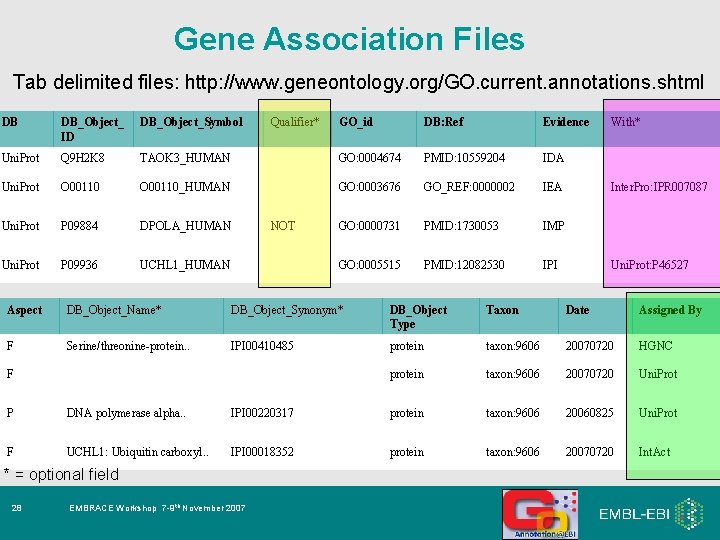
Gene Association Files Tab delimited files: http: //www. geneontology. org/GO. current. annotations. shtml DB DB_Object_ ID DB_Object_Symbol Uni. Prot Q 9 H 2 K 8 Uni. Prot Qualifier* GO_id DB: Ref Evidence TAOK 3_HUMAN GO: 0004674 PMID: 10559204 IDA O 00110_HUMAN GO: 0003676 GO_REF: 0000002 IEA Uni. Prot P 09884 DPOLA_HUMAN GO: 0000731 PMID: 1730053 IMP Uni. Prot P 09936 UCHL 1_HUMAN GO: 0005515 PMID: 12082530 IPI NOT With* Inter. Pro: IPR 007087 Uni. Prot: P 46527 Aspect DB_Object_Name* DB_Object_Synonym* DB_Object Type Taxon Date Assigned By F Serine/threonine-protein. . IPI 00410485 protein taxon: 9606 20070720 HGNC protein taxon: 9606 20070720 Uni. Prot F P DNA polymerase alpha. . IPI 00220317 protein taxon: 9606 20060825 Uni. Prot F UCHL 1: Ubiquitin carboxyl. . IPI 00018352 protein taxon: 9606 20070720 Int. Act * = optional field 28 EMBRACE Workshop 7 -9 th November 2007

http: //www. geneontology. org/GO. current. annotations. shtml 29 EMBRACE Workshop 7 -9 th November 2007

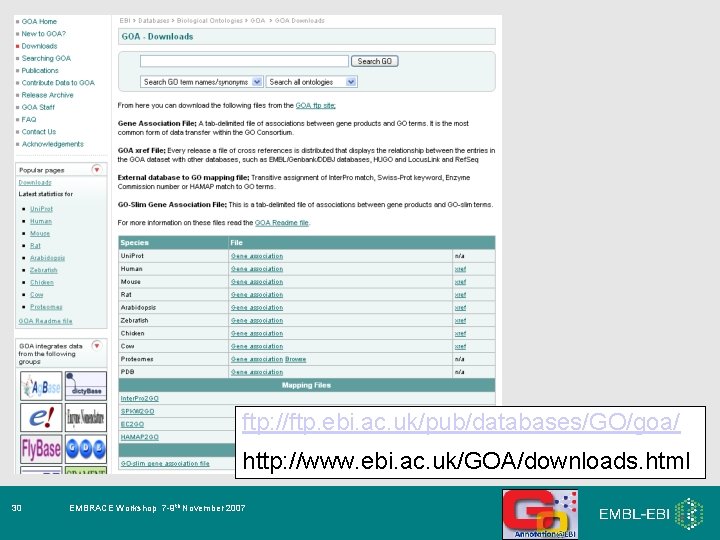
ftp: //ftp. ebi. ac. uk/pub/databases/GO/goa/ http: //www. ebi. ac. uk/GOA/downloads. html 30 EMBRACE Workshop 7 -9 th November 2007

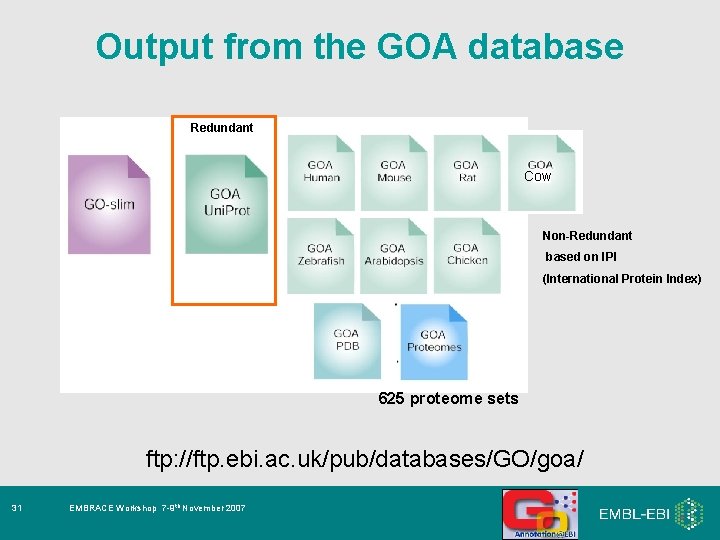
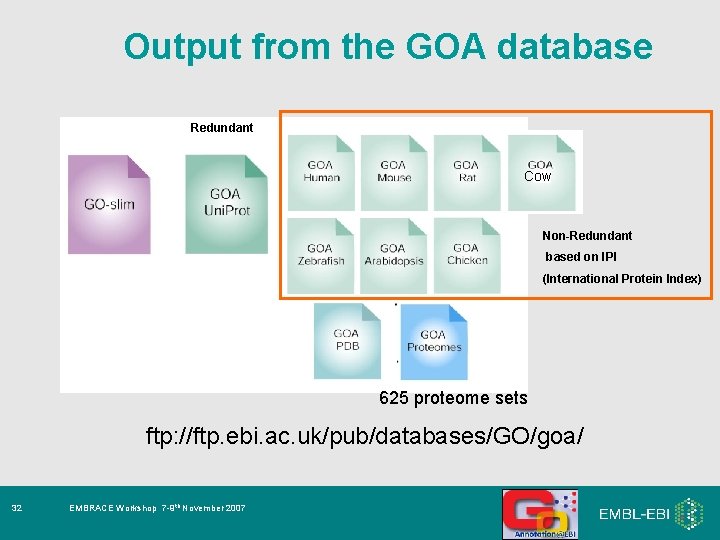
Output from the GOA database Redundant Cow Non-Redundant based on IPI (International Protein Index) 625 proteome sets ftp: //ftp. ebi. ac. uk/pub/databases/GO/goa/ 31 EMBRACE Workshop 7 -9 th November 2007

Output from the GOA database Redundant Cow Non-Redundant based on IPI (International Protein Index) 625 proteome sets ftp: //ftp. ebi. ac. uk/pub/databases/GO/goa/ 32 EMBRACE Workshop 7 -9 th November 2007

… annotations are also displayed in: • All GO Consortium Model Organism Databases integrate and exchange GO annotation data to ensure a comprehensive set of annotations for their organism/area of interest. • Array Products and data analysis Affymetrix Spotfire Almac 33 EMBRACE Workshop 7 -9 th November 2007

… and Numerous Third Party Tools (http: //www. geneontology. org/GO. tools. shtml) 34 EMBRACE Workshop 7 -9 th November 2007

What’s new on the GO annotation front? 35 EMBRACE Workshop 7 -9 th November 2007

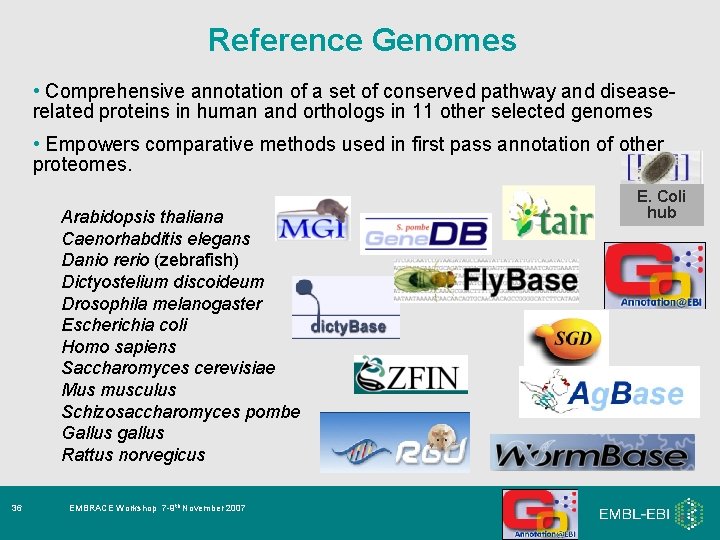
Reference Genomes • Comprehensive annotation of a set of conserved pathway and diseaserelated proteins in human and orthologs in 11 other selected genomes • Empowers comparative methods used in first pass annotation of other proteomes. Arabidopsis thaliana Caenorhabditis elegans Danio rerio (zebrafish) Dictyostelium discoideum Drosophila melanogaster Escherichia coli Homo sapiens Saccharomyces cerevisiae Mus musculus Schizosaccharomyces pombe Gallus gallus Rattus norvegicus 36 EMBRACE Workshop 7 -9 th November 2007 E. Coli hub

GOA annotation focuses Cardiovascular GO annotation Grant with the British Heart Foundation to support a collaboration with HGNC curators to provide full Gene Ontology annotation to genes associated with cardiovascular processes wiki: http: //wiki. geneontology. org/index. php/Cardiovascular Immune GO annotation Interest in actively GO annotating immune relevant genes. GOA, UCL and MGI are collaborating to improve annotation for immunologically-important genes, WT grant pending. wiki: http: //wiki. geneontology. org/index. php/Immunology 37 EMBRACE Workshop 7 -9 th November 2007

Electronic Annotation developments New mappings: • Swiss-Prot Subcellar Location to GO (just released) • Swiss-Prot Uni. Pathway Expansion of existing methods • Ensembl Compara species expansion 38 EMBRACE Workshop 7 -9 th November 2007

Acknowledgements Rolf Apweiler. Head of the EBI protein sequence database group Emily Dimmer Evelyn Camon Rachael Huntley Daniel Barrell David Binns Contact the GOA team: GOA web page: 39 goa@ebi. ac. uk http: //www. ebi. ac. uk/goa The Gene Ontology Consortium and 1. 5 members of GOA currently supported by an P 41 grant from the National Human Genome Research Institute (NHGRI) [grant HG 002273], GOA is also supported by core EMBL funding and BBSRC Tools and th November EMBRACE Workshop 7 -9 2007 Resources grant.