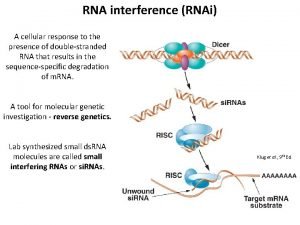
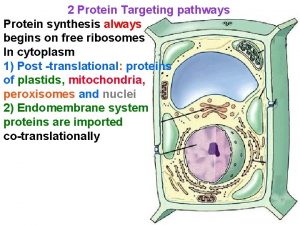
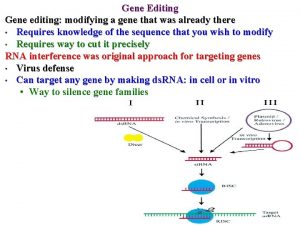
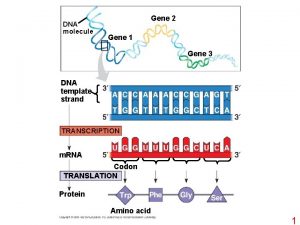
Gene Targeting GT RNAi Gene targeting GT refers








- Slides: 21

Gene Targeting ● ● GT是指在基因组的原位实现对基因的定点突变、定 点整合、基因置换及基因修复等。对基因功能的研 究提供了极大的方便,比基因的反义抑制和RNAi更 直观可靠,尤其在鉴定基因家族成员的功能时更是 如此。 Gene targeting (GT) refers to the alteration of a specific DNA sequence in an endogenous gene at its original locus in the genome by homologous recombination (HR) and, often, to the conversion of the endogenous gene into a designed sequence, i. e. base changes or gene disruptions through gene replacements.

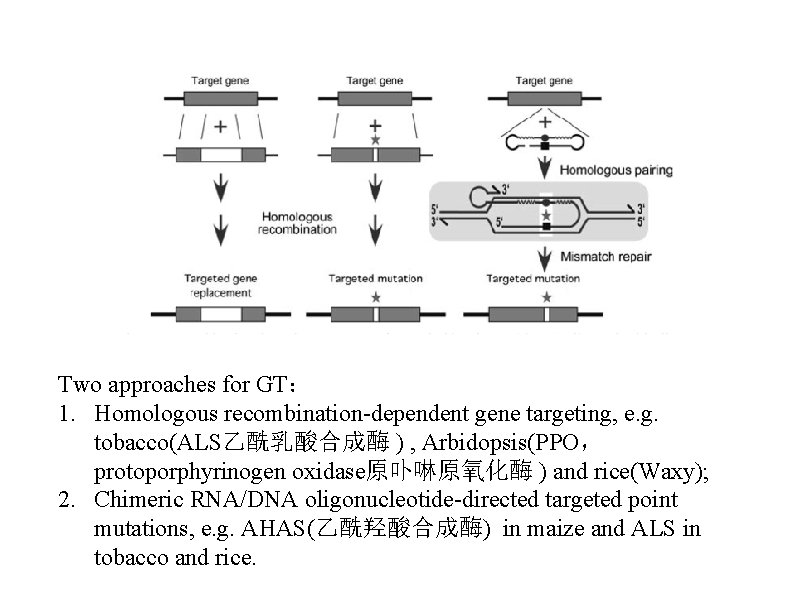
Two approaches for GT: 1. Homologous recombination-dependent gene targeting, e. g. tobacco(ALS乙酰乳酸合成酶 ) , Arbidopsis(PPO, protoporphyrinogen oxidase原卟啉原氧化酶 ) and rice(Waxy); 2. Chimeric RNA/DNA oligonucleotide-directed targeted point mutations, e. g. AHAS(乙酰羟酸合成酶) in maize and ALS in tobacco and rice.



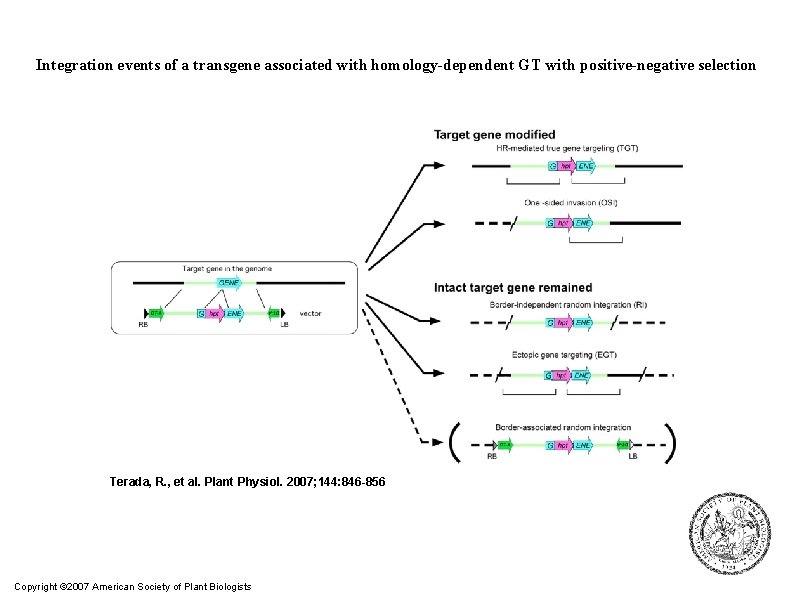
Integration events of a transgene associated with homology-dependent GT with positive-negative selection Terada, R. , et al. Plant Physiol. 2007; 144: 846 -856 Copyright © 2007 American Society of Plant Biologists

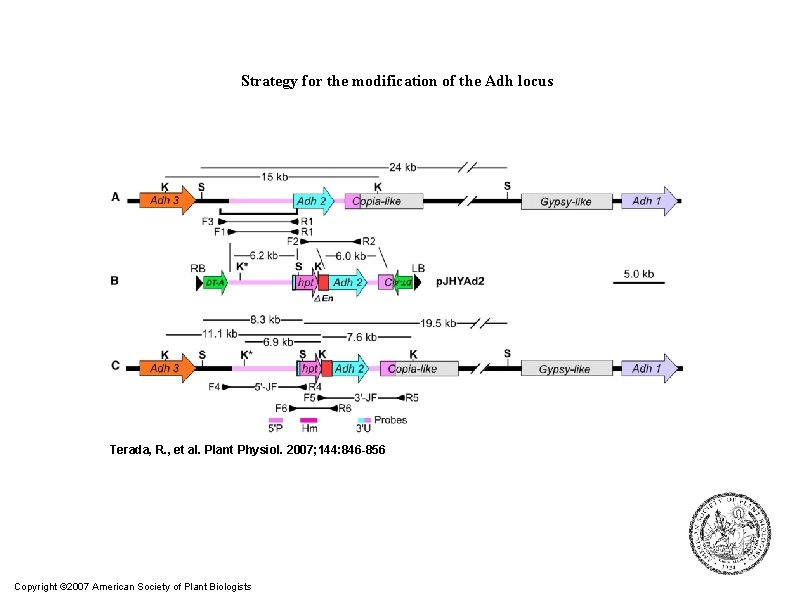
Strategy for the modification of the Adh locus Terada, R. , et al. Plant Physiol. 2007; 144: 846 -856 Copyright © 2007 American Society of Plant Biologists


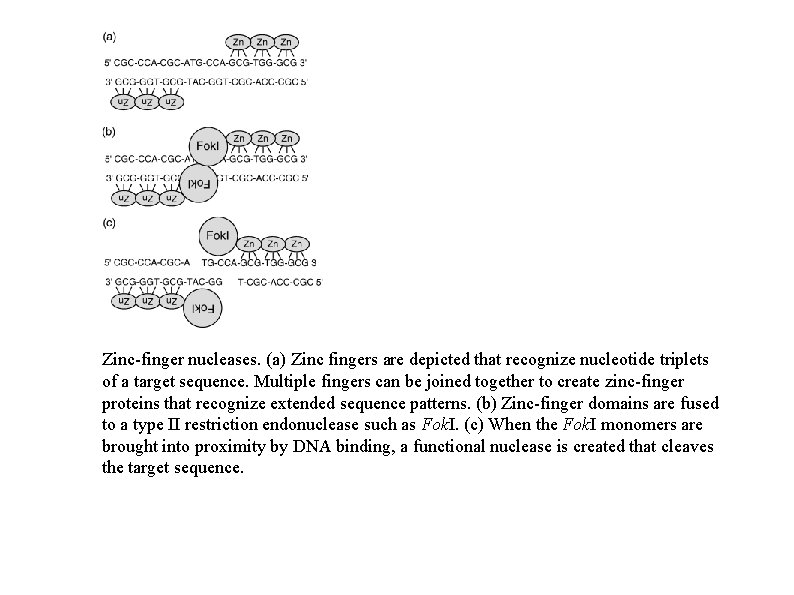
Zinc-finger nucleases. (a) Zinc fingers are depicted that recognize nucleotide triplets of a target sequence. Multiple fingers can be joined together to create zinc-finger proteins that recognize extended sequence patterns. (b) Zinc-finger domains are fused to a type II restriction endonuclease such as Fok. I. (c) When the Fok. I monomers are brought into proximity by DNA binding, a functional nuclease is created that cleaves the target sequence.

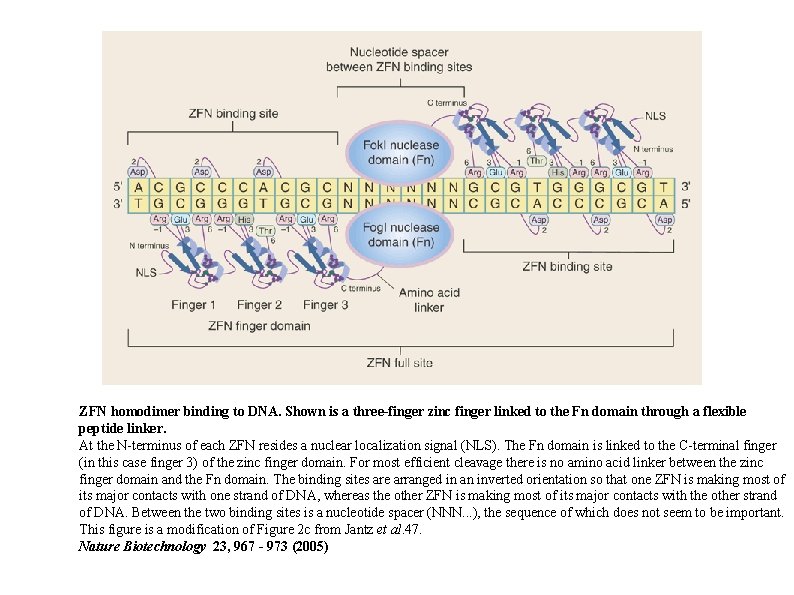
ZFN homodimer binding to DNA. Shown is a three-finger zinc finger linked to the Fn domain through a flexible peptide linker. At the N-terminus of each ZFN resides a nuclear localization signal (NLS). The Fn domain is linked to the C-terminal finger (in this case finger 3) of the zinc finger domain. For most efficient cleavage there is no amino acid linker between the zinc finger domain and the Fn domain. The binding sites are arranged in an inverted orientation so that one ZFN is making most of its major contacts with one strand of DNA, whereas the other ZFN is making most of its major contacts with the other strand of DNA. Between the two binding sites is a nucleotide spacer (NNN. . . ), the sequence of which does not seem to be important. This figure is a modification of Figure 2 c from Jantz et al. 47. Nature Biotechnology 23, 967 - 973 (2005)

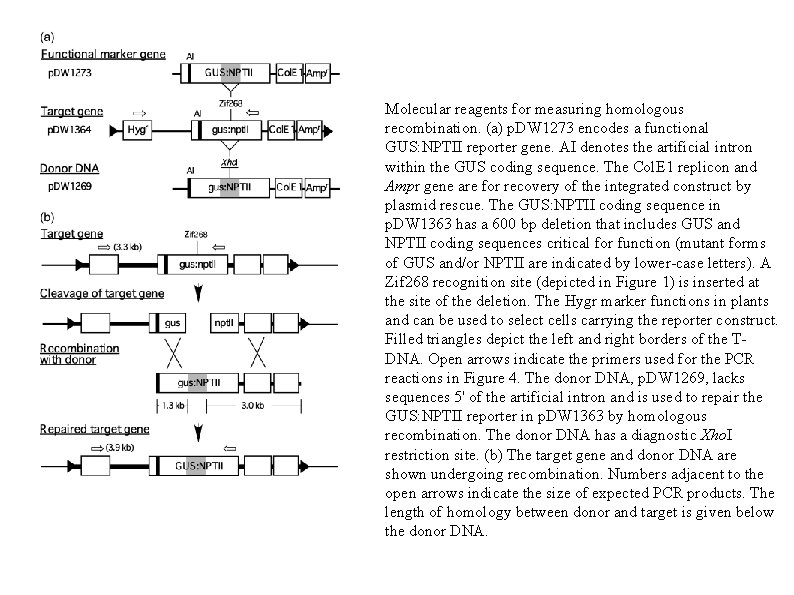
Molecular reagents for measuring homologous recombination. (a) p. DW 1273 encodes a functional GUS: NPTII reporter gene. AI denotes the artificial intron within the GUS coding sequence. The Col. E 1 replicon and Ampr gene are for recovery of the integrated construct by plasmid rescue. The GUS: NPTII coding sequence in p. DW 1363 has a 600 bp deletion that includes GUS and NPTII coding sequences critical for function (mutant forms of GUS and/or NPTII are indicated by lower-case letters). A Zif 268 recognition site (depicted in Figure 1) is inserted at the site of the deletion. The Hygr marker functions in plants and can be used to select cells carrying the reporter construct. Filled triangles depict the left and right borders of the TDNA. Open arrows indicate the primers used for the PCR reactions in Figure 4. The donor DNA, p. DW 1269, lacks sequences 5' of the artificial intron and is used to repair the GUS: NPTII reporter in p. DW 1363 by homologous recombination. The donor DNA has a diagnostic Xho. I restriction site. (b) The target gene and donor DNA are shown undergoing recombination. Numbers adjacent to the open arrows indicate the size of expected PCR products. The length of homology between donor and target is given below the donor DNA.

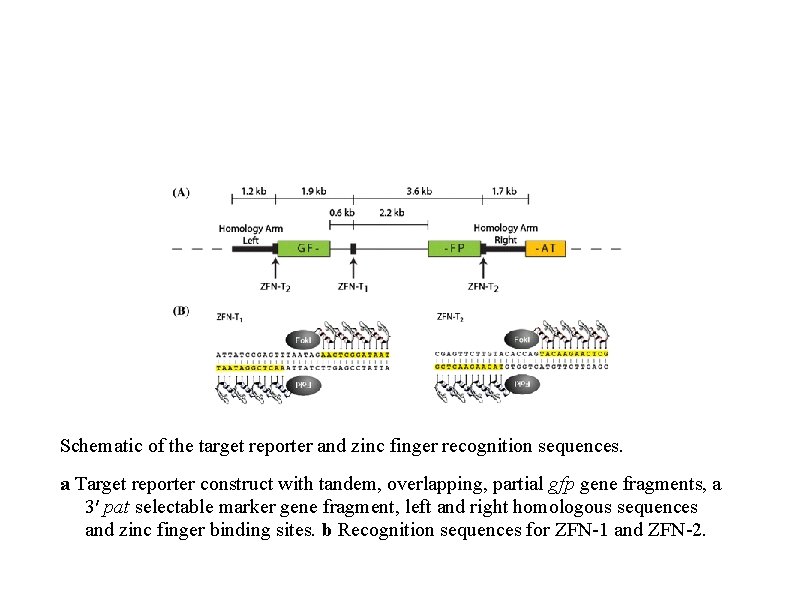
Schematic of the target reporter and zinc finger recognition sequences. a Target reporter construct with tandem, overlapping, partial gfp gene fragments, a 3′ pat selectable marker gene fragment, left and right homologous sequences and zinc finger binding sites. b Recognition sequences for ZFN-1 and ZFN-2.

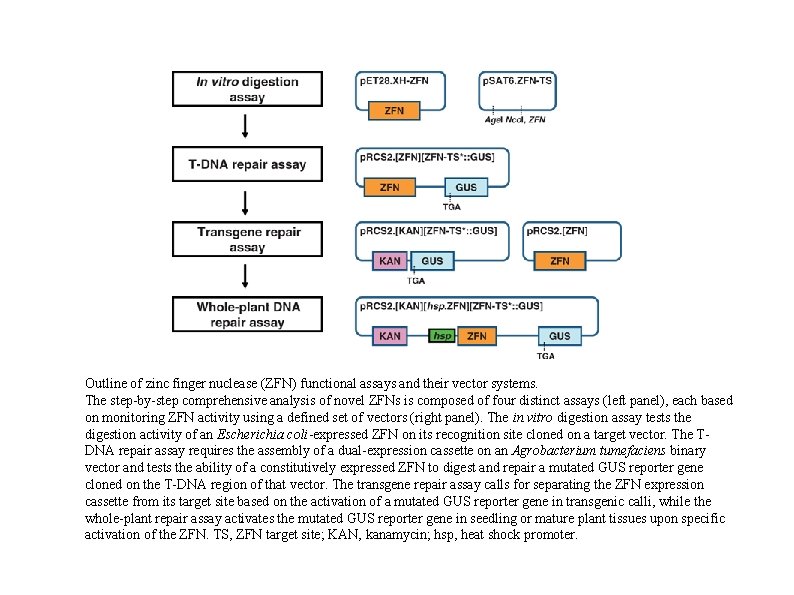
Outline of zinc finger nuclease (ZFN) functional assays and their vector systems. The step-by-step comprehensive analysis of novel ZFNs is composed of four distinct assays (left panel), each based on monitoring ZFN activity using a defined set of vectors (right panel). The in vitro digestion assay tests the digestion activity of an Escherichia coli-expressed ZFN on its recognition site cloned on a target vector. The TDNA repair assay requires the assembly of a dual-expression cassette on an Agrobacterium tumefaciens binary vector and tests the ability of a constitutively expressed ZFN to digest and repair a mutated GUS reporter gene cloned on the T-DNA region of that vector. The transgene repair assay calls for separating the ZFN expression cassette from its target site based on the activation of a mutated GUS reporter gene in transgenic calli, while the whole-plant repair assay activates the mutated GUS reporter gene in seedling or mature plant tissues upon specific activation of the ZFN. TS, ZFN target site; KAN, kanamycin; hsp, heat shock promoter.


● Structural features of the zinc finger nuclease (ZFN) assembly and expression vector systems. A zinc finger protein (ZFP) coding sequence can be assembled by Klenow/PCR using a combination of overlapping backbone and sequence-dependent oligonucleotides fused to the Fok. I endonuclease domain in p. SAT 4. 35 SP. NLSFok. I, producing the plant expression vector p. SAT 4. 35 SP. ZFN. The entire ZFN coding sequence can be transferred onto a p. ET 28. XH-based vector producing a p. ET 28. XH-ZFN vector, suitable for ZFN expression in bacterial cells and for in vivo digestion assays. The p. SAT 4. 35 SP. ZFN can be modified by replacing the 35 S constitutive promoter with a heat-shock-inducible promoter, producing a plasmid that is useful for the whole-plant DNA repair assay. A plant-selectable marker, a ZFN and a mutated GUS reporter expression cassette can be mounted onto a p. RCS 2 -based binary plant transformation vector using a combination of rare-cutting restriction enzymes and can then be used for various in planta assays. 35 SP, 35 S promoter; 35 ST, 35 S terminator; hsp, heat shock promoter.

Delivery of Multiple Transgenes to Plant Cells ● The growing interest in dissecting and analyzing complex metabolic pathways and the need to exploit the full potential of multigene traits for plant biotechnology mandate the development of new methods and tools for the delivery and stacking of multiple genes in plant cells.

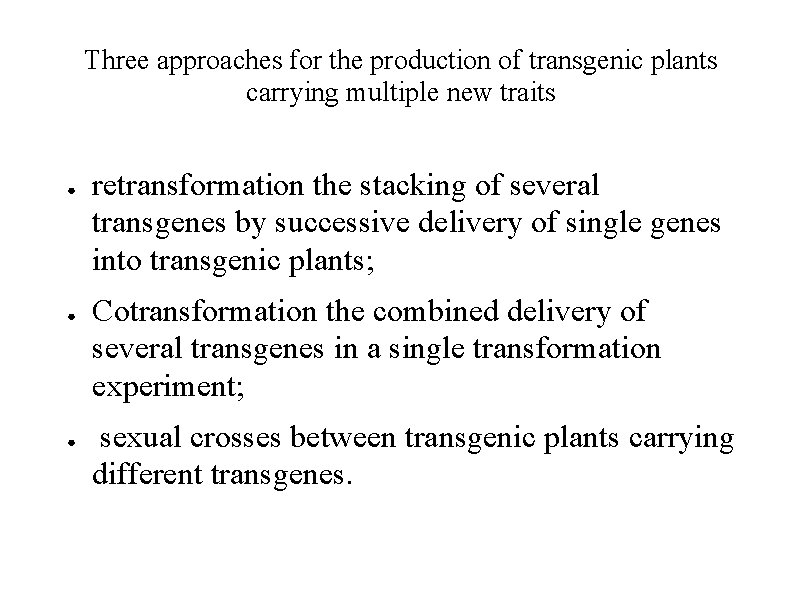
Three approaches for the production of transgenic plants carrying multiple new traits ● ● ● retransformation the stacking of several transgenes by successive delivery of single genes into transgenic plants; Cotransformation the combined delivery of several transgenes in a single transformation experiment; sexual crosses between transgenic plants carrying different transgenes.

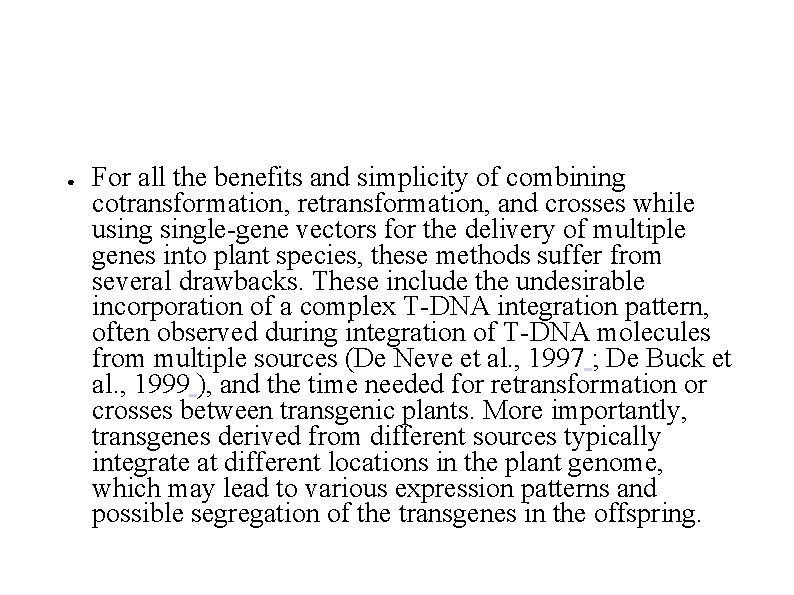
● For all the benefits and simplicity of combining cotransformation, retransformation, and crosses while usingle-gene vectors for the delivery of multiple genes into plant species, these methods suffer from several drawbacks. These include the undesirable incorporation of a complex T-DNA integration pattern, often observed during integration of T-DNA molecules from multiple sources (De Neve et al. , 1997 ; De Buck et al. , 1999 ), and the time needed for retransformation or crosses between transgenic plants. More importantly, transgenes derived from different sources typically integrate at different locations in the plant genome, which may lead to various expression patterns and possible segregation of the transgenes in the offspring.

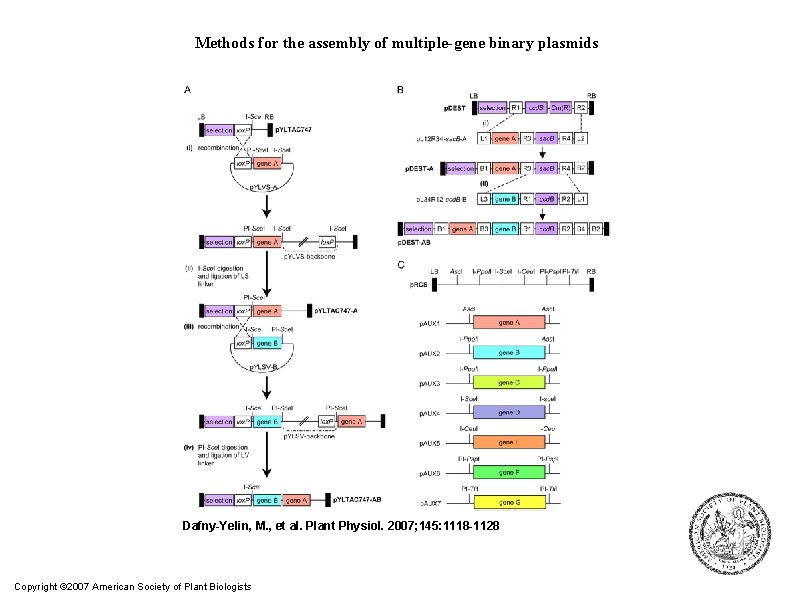
● Methods for the assembly of multiple-gene binary plasmids. A, The Cre/lox. Pmediated multigene assembly process. i, Cre/lox. P recombination of the p. YLVS -A plasmid into the p. YLTAC 747 acceptor binary plasmid. ii, Release of the p. YLVS backbone by I-Sce. I digestion and ligation with a LS linker. This ligation abolishes the I-Sce. I site from p. YLTAC 747 -A. iii, Cre/lox. P recombination of the p. YLSV-B plasmid into the p. YLTAC 747 -A and release of the p. YLSV backbone by PI-Sce. I digestion and ligation with a LV linker (iv). B, The Multi. Round Gateway assembly process. i, Gateway recombination between att. L 1 and att. R 1 and between att. L 1 and att. R 1 sites by LR clonase and conversion of the ccd. B-based binary p. DEST vector into a sac. B-based Destination vector. ii, Gateway recombination between att. L 3 and att. R 3 and between att. L 4 and att. R 4 sites by LR clonase and reconversion of the destination binary vector into a ccd. B-based plasmid. C, The homing endonucleases p. RCS/p. AUX vector system. Assembly of a multigene binary plasmid is achieved by successive cloning of various gene expression cassettes using rare-cutting homing endonucleases.

Methods for the assembly of multiple-gene binary plasmids Dafny-Yelin, M. , et al. Plant Physiol. 2007; 145: 1118 -1128 Copyright © 2007 American Society of Plant Biologists


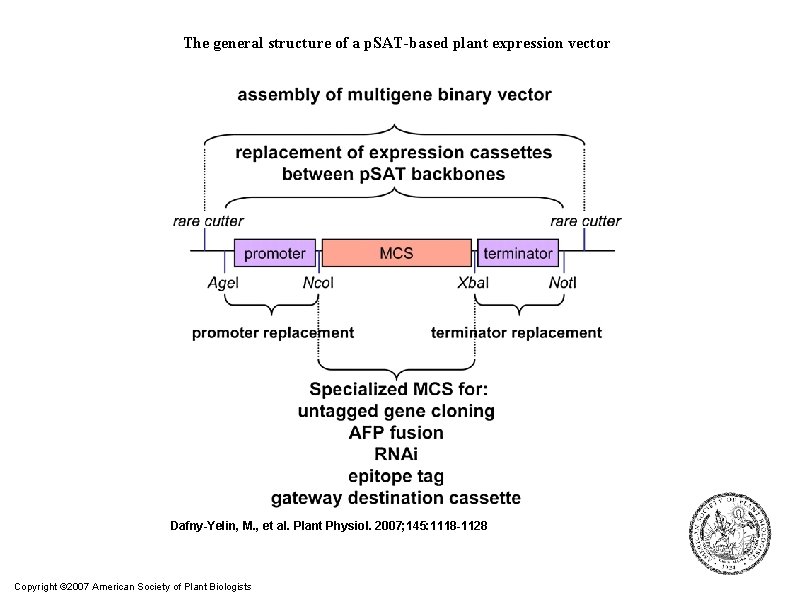
The general structure of a p. SAT-based plant expression vector Dafny-Yelin, M. , et al. Plant Physiol. 2007; 145: 1118 -1128 Copyright © 2007 American Society of Plant Biologists
 Ahringer rnai library
Ahringer rnai library Rnai
Rnai Gene by gene test results
Gene by gene test results Chapter 17: from gene to protein
Chapter 17: from gene to protein Connection targeting
Connection targeting Resume jurnal
Resume jurnal Mktg 8
Mktg 8 Bssu
Bssu Protien sorting
Protien sorting Gdn targeting options
Gdn targeting options Presciber segmentation
Presciber segmentation Find-fix-finish-exploit-analyze targeting model
Find-fix-finish-exploit-analyze targeting model Differentiated undifferentiated and concentrated marketing
Differentiated undifferentiated and concentrated marketing Nivea positioning strategy
Nivea positioning strategy Marketing differenziato
Marketing differenziato Psychographic segmentation of nivea
Psychographic segmentation of nivea Sony market segmentation, targeting and positioning
Sony market segmentation, targeting and positioning Concentrated targeting
Concentrated targeting Global segmentation targeting and positioning
Global segmentation targeting and positioning Definisi targeting
Definisi targeting Bases of market segmentation
Bases of market segmentation Stp of colgate
Stp of colgate