Exome sequencing and characterization of 49 960 individuals


- Slides: 19

Exome sequencing and characterization of 49, 960 individuals in the UK Biobank Van Hout et al, Oct 2020 �Genotyping, imputation & sequencing �Whole-Exome Sequencing ◦ What is the ‘exome’? ◦ My experience �UKB 50 k Exomes – “flagship” paper Genetics Forum: 05/11/20 Mesut Erzurumluoglu 1

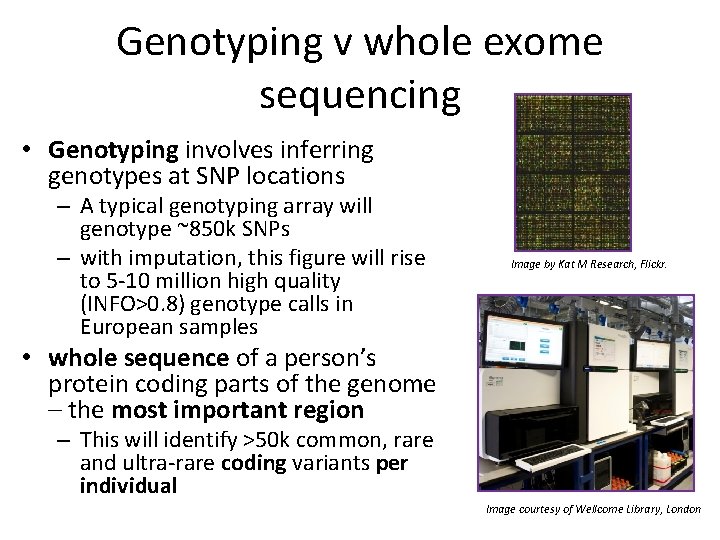
Genotyping v whole exome sequencing • Genotyping involves inferring genotypes at SNP locations – A typical genotyping array will genotype ~850 k SNPs – with imputation, this figure will rise to 5 -10 million high quality (INFO>0. 8) genotype calls in European samples Image by Kat M Research, Flickr. • whole sequence of a person’s protein coding parts of the genome – the most important region – This will identify >50 k common, rare and ultra-rare coding variants per individual Image courtesy of Wellcome Library, London

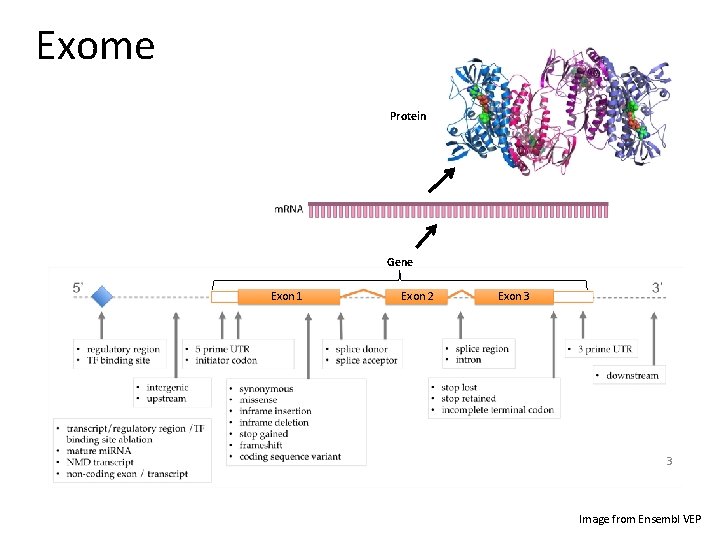
Exome Protein Gene Exon 1 Exon 2 Exon 3 3 Image from Ensembl VEP

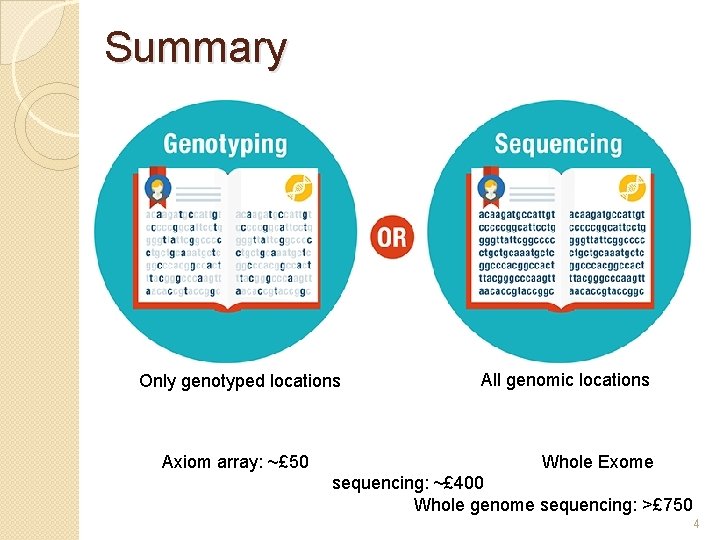
Summary Only genotyped locations Axiom array: ~£ 50 All genomic locations Whole Exome sequencing: ~£ 400 Whole genome sequencing: >£ 750 4

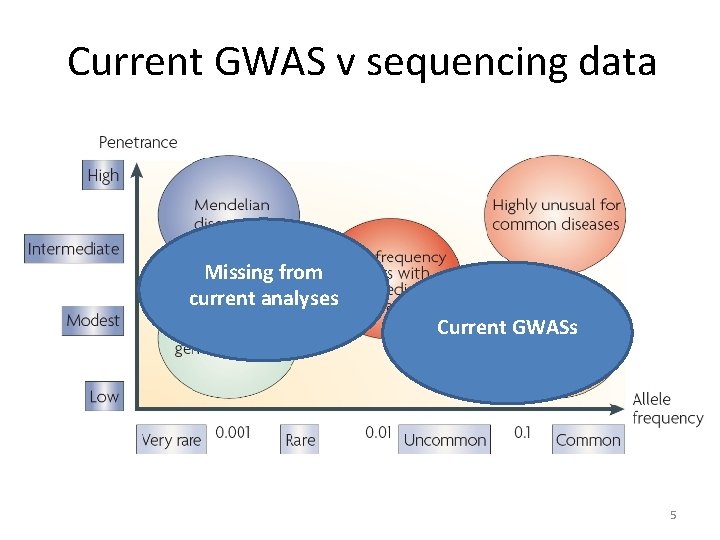
Current GWAS v sequencing data Missing from current analyses Current GWASs 5

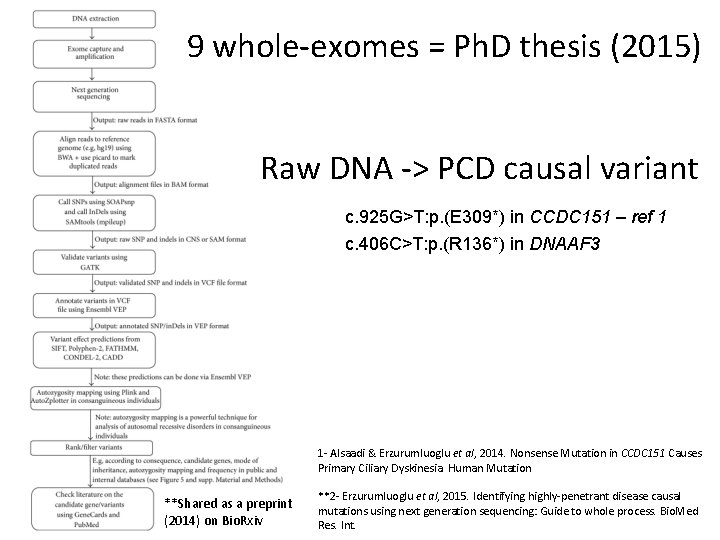
9 whole-exomes = Ph. D thesis (2015) Raw DNA -> PCD causal variant c. 925 G>T: p. (E 309*) in CCDC 151 – ref 1 c. 406 C>T: p. (R 136*) in DNAAF 3 1 - Alsaadi & Erzurumluoglu et al, 2014. Nonsense Mutation in CCDC 151 Causes Primary Ciliary Dyskinesia. Human Mutation **Shared as a preprint (2014) on Bio. Rxiv **2 - Erzurumluoglu et al, 2015. Identifying highly-penetrant disease causal mutations using next generation sequencing: Guide to whole process. Bio. Med Res. Int.

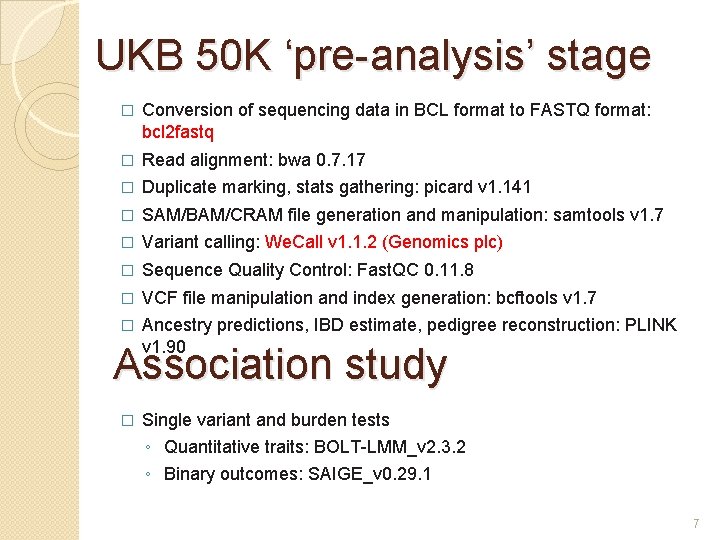
UKB 50 K ‘pre-analysis’ stage � Conversion of sequencing data in BCL format to FASTQ format: bcl 2 fastq � Read alignment: bwa 0. 7. 17 � Duplicate marking, stats gathering: picard v 1. 141 � SAM/BAM/CRAM file generation and manipulation: samtools v 1. 7 � Variant calling: We. Call v 1. 1. 2 (Genomics plc) � Sequence Quality Control: Fast. QC 0. 11. 8 � VCF file manipulation and index generation: bcftools v 1. 7 � Ancestry predictions, IBD estimate, pedigree reconstruction: PLINK v 1. 90 Association study � Single variant and burden tests ◦ Quantitative traits: BOLT-LMM_v 2. 3. 2 ◦ Binary outcomes: SAIGE_v 0. 29. 1 7

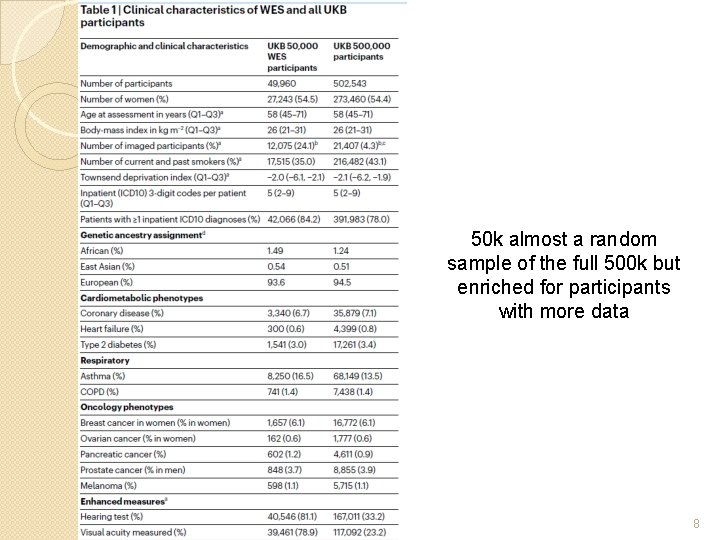
50 k almost a random sample of the full 500 k but enriched for participants with more data 8

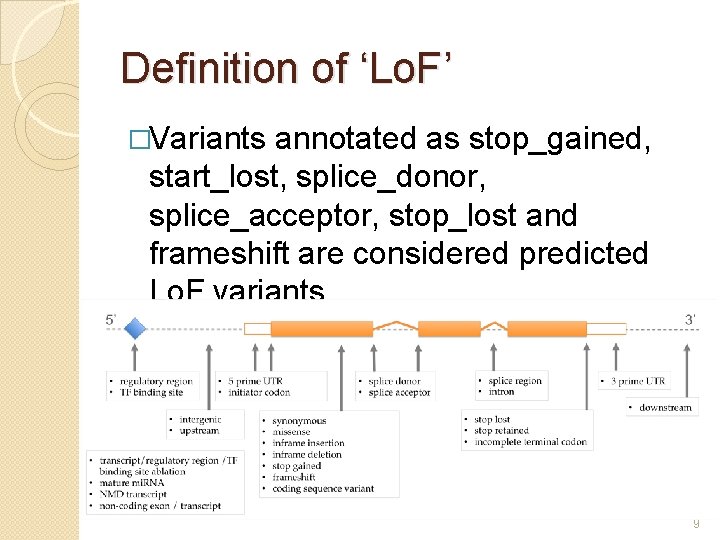
Definition of ‘Lo. F’ �Variants annotated as stop_gained, start_lost, splice_donor, splice_acceptor, stop_lost and frameshift are considered predicted Lo. F variants 9

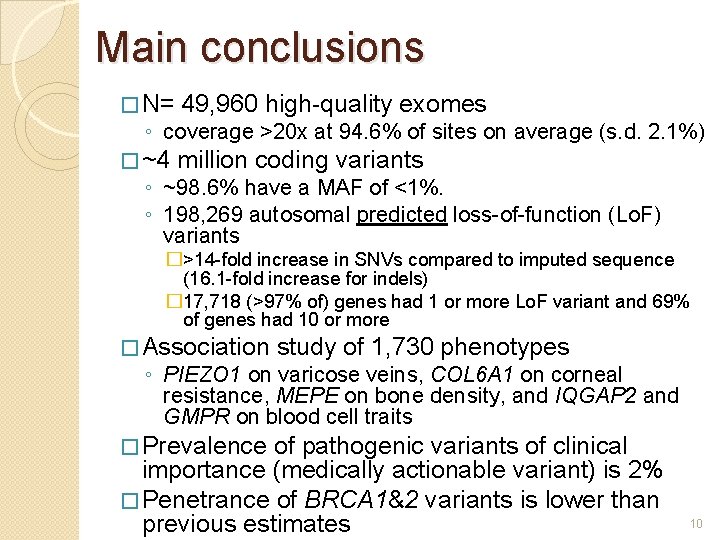
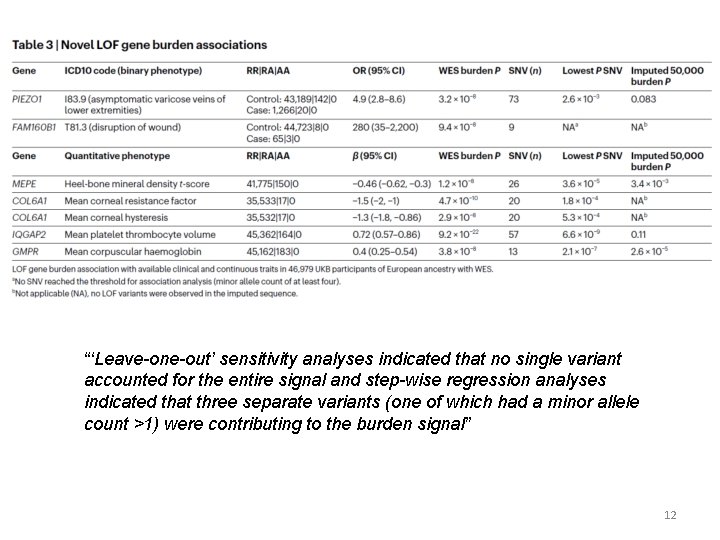
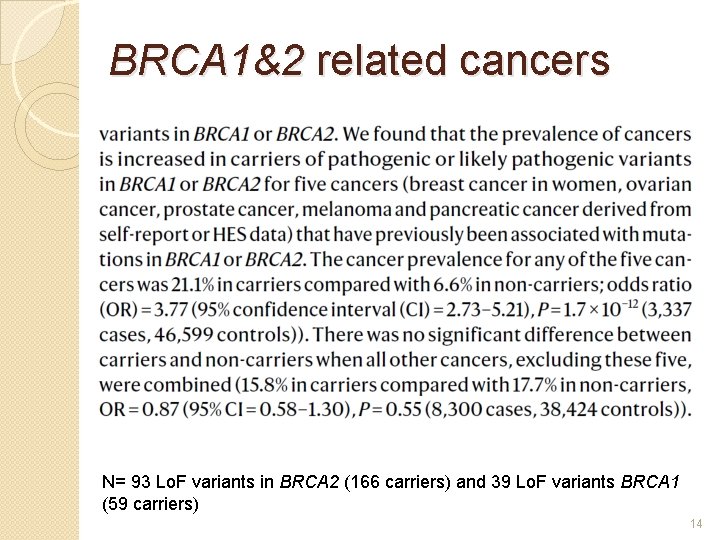
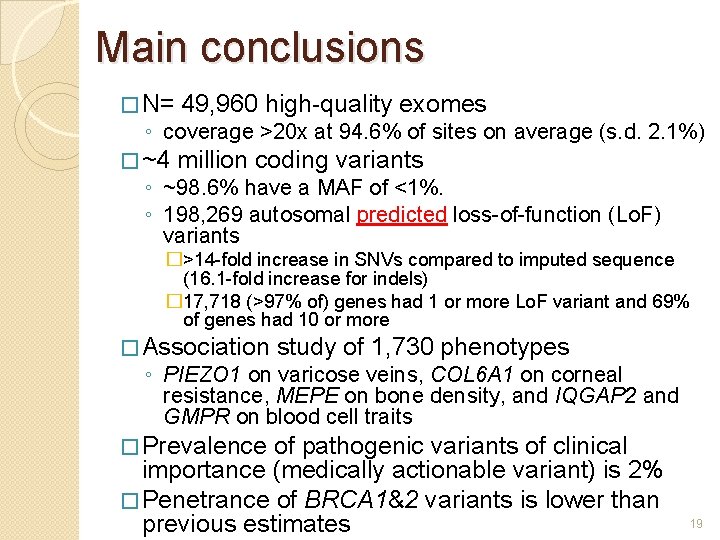
Main conclusions � N= 49, 960 high-quality exomes � ~4 million coding variants ◦ coverage >20 x at 94. 6% of sites on average (s. d. 2. 1%) ◦ ~98. 6% have a MAF of <1%. ◦ 198, 269 autosomal predicted loss-of-function (Lo. F) variants �>14 -fold increase in SNVs compared to imputed sequence (16. 1 -fold increase for indels) � 17, 718 (>97% of) genes had 1 or more Lo. F variant and 69% of genes had 10 or more � Association study of 1, 730 phenotypes ◦ PIEZO 1 on varicose veins, COL 6 A 1 on corneal resistance, MEPE on bone density, and IQGAP 2 and GMPR on blood cell traits � Prevalence of pathogenic variants of clinical importance (medically actionable variant) is 2% � Penetrance of BRCA 1&2 variants is lower than previous estimates 10

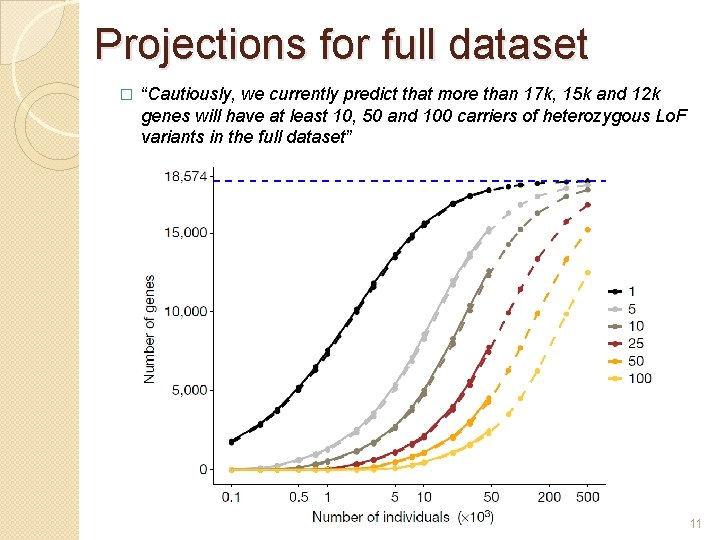
Projections for full dataset � “Cautiously, we currently predict that more than 17 k, 15 k and 12 k genes will have at least 10, 50 and 100 carriers of heterozygous Lo. F variants in the full dataset” 11

“‘Leave-one-out’ sensitivity analyses indicated that no single variant accounted for the entire signal and step-wise regression analyses indicated that three separate variants (one of which had a minor allele count >1) were contributing to the burden signal” 12


BRCA 1&2 related cancers N= 93 Lo. F variants in BRCA 2 (166 carriers) and 39 Lo. F variants BRCA 1 (59 carriers) 14

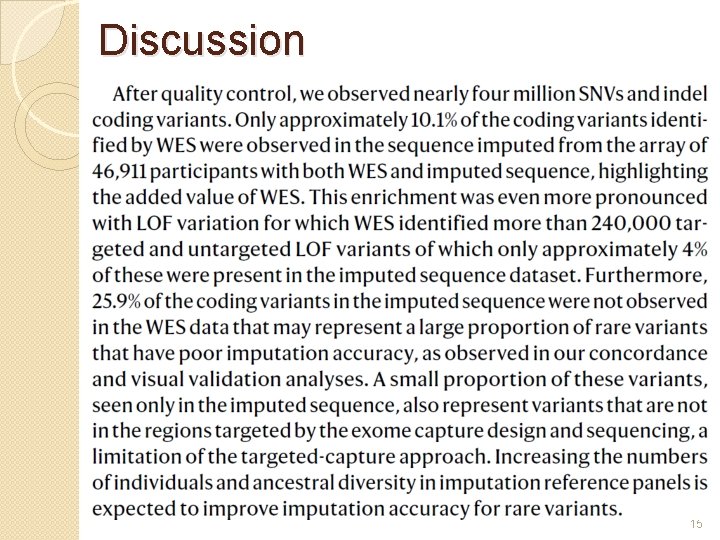
Discussion 15

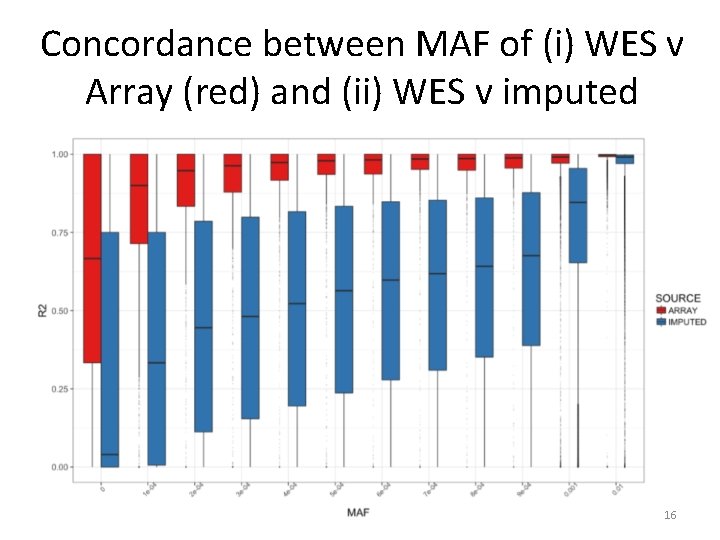
Concordance between MAF of (i) WES v Array (red) and (ii) WES v imputed 16

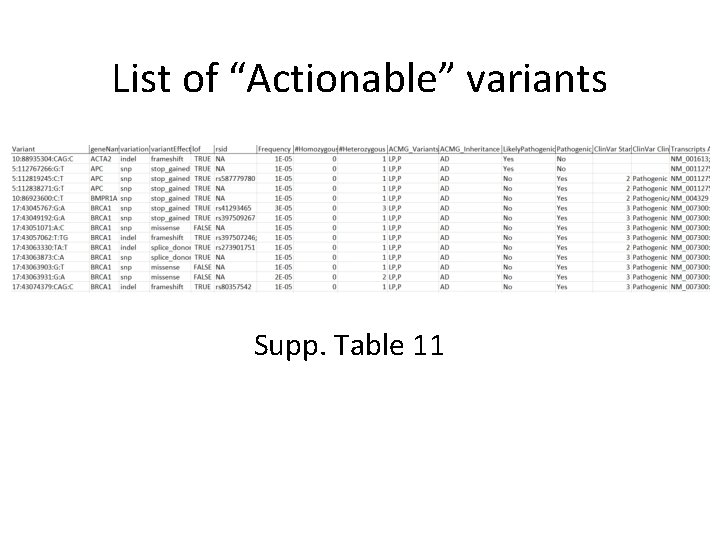
List of “Actionable” variants Supp. Table 11

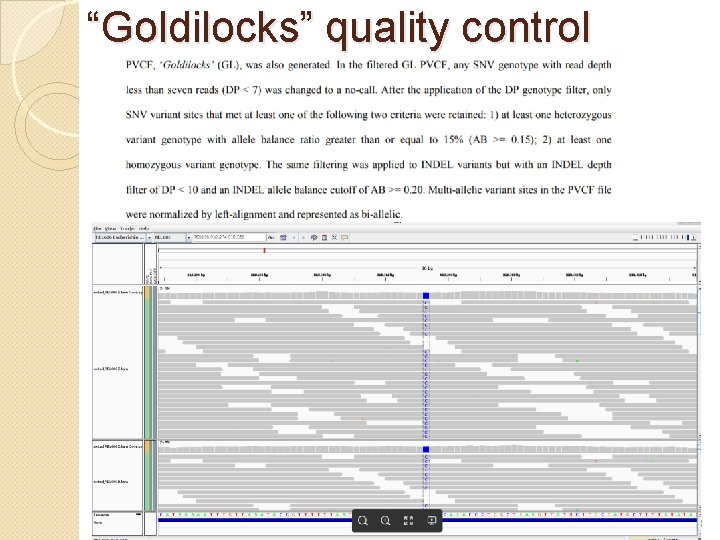
“Goldilocks” quality control 18

Main conclusions � N= 49, 960 high-quality exomes � ~4 million coding variants ◦ coverage >20 x at 94. 6% of sites on average (s. d. 2. 1%) ◦ ~98. 6% have a MAF of <1%. ◦ 198, 269 autosomal predicted loss-of-function (Lo. F) variants �>14 -fold increase in SNVs compared to imputed sequence (16. 1 -fold increase for indels) � 17, 718 (>97% of) genes had 1 or more Lo. F variant and 69% of genes had 10 or more � Association study of 1, 730 phenotypes ◦ PIEZO 1 on varicose veins, COL 6 A 1 on corneal resistance, MEPE on bone density, and IQGAP 2 and GMPR on blood cell traits � Prevalence of pathogenic variants of clinical importance (medically actionable variant) is 2% � Penetrance of BRCA 1&2 variants is lower than previous estimates 19
 Exome sequencing project
Exome sequencing project Exome
Exome Khan academy psat scores
Khan academy psat scores Horiba la950
Horiba la950 Atm-960
Atm-960 1960 sonrası türk hikayesinin genel özellikleri
1960 sonrası türk hikayesinin genel özellikleri 9 596 960
9 596 960 Cina superficie km2
Cina superficie km2 Indirect characterization definition
Indirect characterization definition What is direct characterization?
What is direct characterization? Sequence, selection, and iteration
Sequence, selection, and iteration Microprogrammed control unit
Microprogrammed control unit Scheduling rules operations management
Scheduling rules operations management Basil khuder
Basil khuder Sequential conditional and iterative
Sequential conditional and iterative Get sequence get another sequence pseudocode
Get sequence get another sequence pseudocode Sequencing strategies and tactics
Sequencing strategies and tactics Cloning and sequencing explorer series
Cloning and sequencing explorer series Sequencing batch reactor advantages and disadvantages
Sequencing batch reactor advantages and disadvantages Risk management for enterprises and individuals
Risk management for enterprises and individuals