Exome Sequencing as Molecular Diagnostic Tool of Mendelian











- Slides: 38

Exome Sequencing as Molecular Diagnostic Tool of Mendelian Diseases BIOS 6660 Hung-Chun (James) Yu Shaikh Lab 04/28/2014

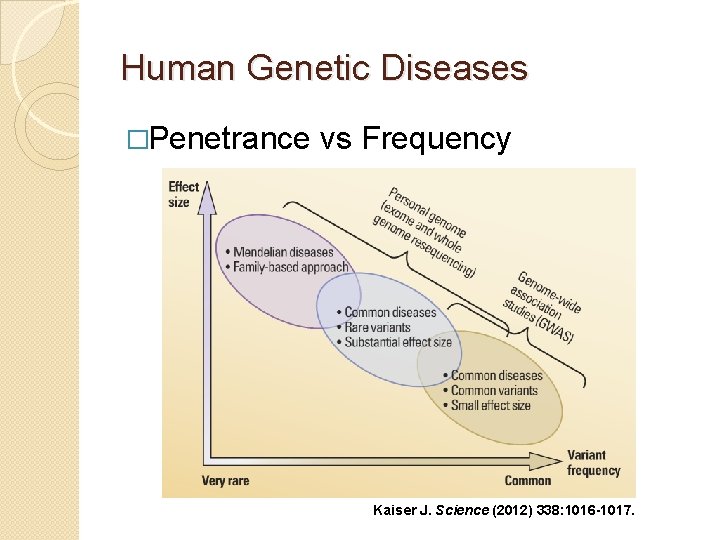
Human Genetic Diseases �Penetrance vs Frequency Kaiser J. Science (2012) 338: 1016 -1017.

Human Genetic Diseases �Complex • • Disorder Polygenic, many genes. Low penetrance/effect size. Multifactorial, environmental, dietary. Examples: heart disease, diabetes, obesity, autism, etc. �Mendelian • • • Disorder Monogenic or polygenic. Full or high penetrance/effect size. Examples: sickle cell anemia and cystic fibrosis.

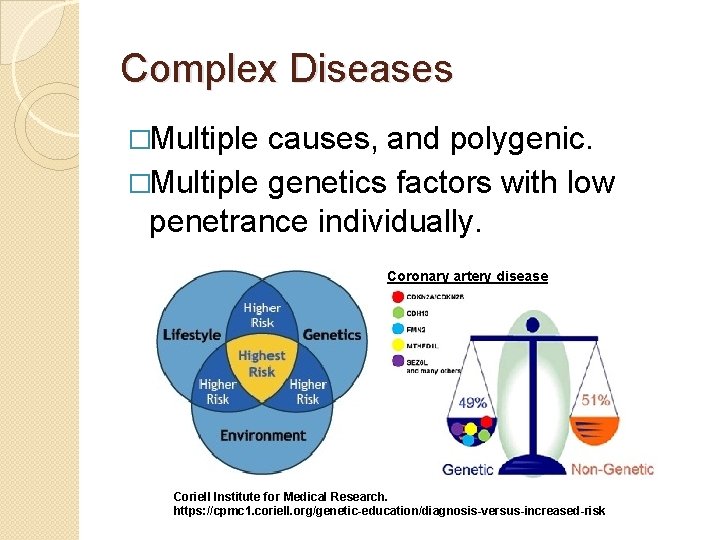
Complex Diseases �Multiple causes, and polygenic. �Multiple genetics factors with low penetrance individually. Coronary artery disease Coriell Institute for Medical Research. https: //cpmc 1. coriell. org/genetic-education/diagnosis-versus-increased-risk

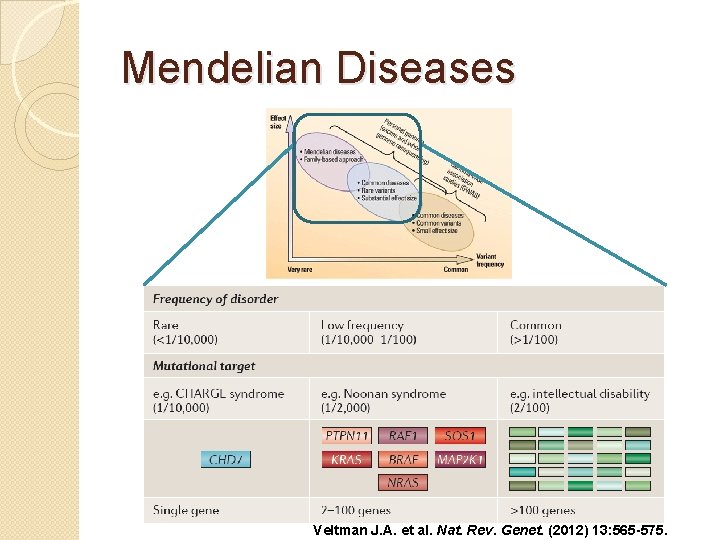
Mendelian Diseases Veltman J. A. et al. Nat. Rev. Genet. (2012) 13: 565 -575.

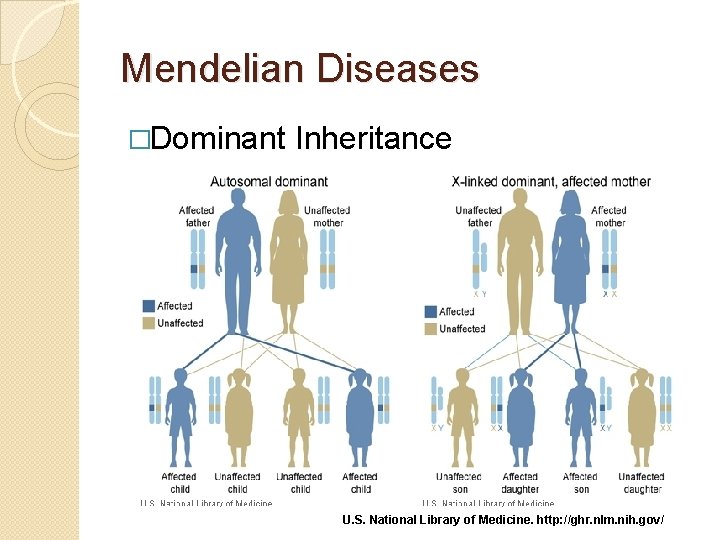
Mendelian Diseases �Dominant Inheritance U. S. National Library of Medicine. http: //ghr. nlm. nih. gov/

Mendelian Diseases �Recessive Inheritance U. S. National Library of Medicine. http: //ghr. nlm. nih. gov/

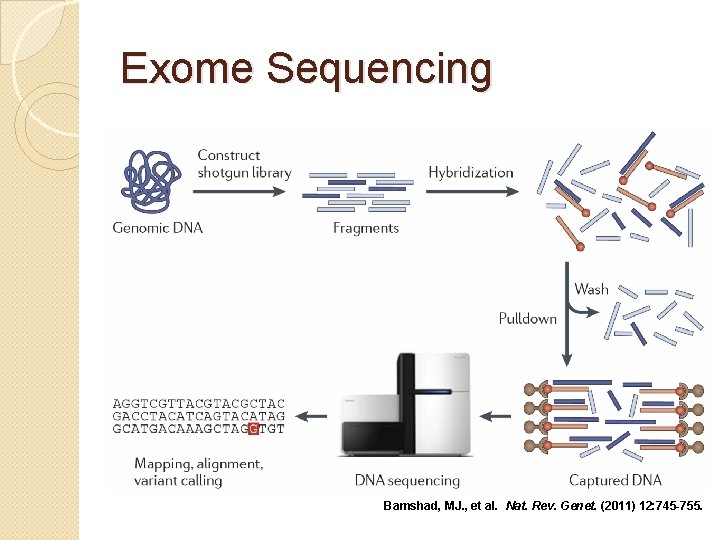
Exome Sequencing Bamshad, MJ. , et al. Nat. Rev. Genet. (2011) 12: 745 -755.

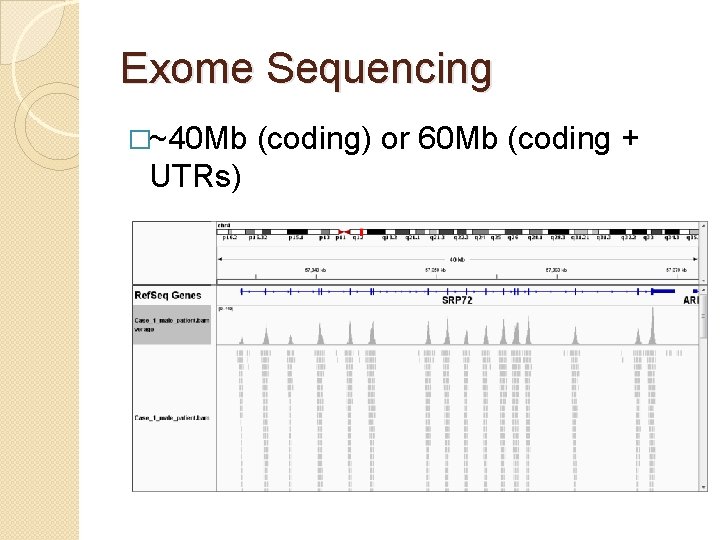
Exome Sequencing �~40 Mb UTRs) (coding) or 60 Mb (coding +

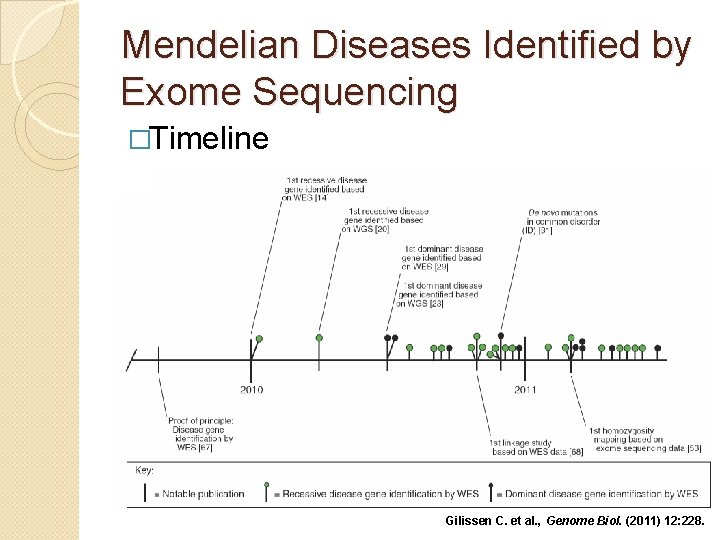
Mendelian Diseases Identified by Exome Sequencing �Timeline Gilissen C. et al. , Genome Biol. (2011) 12: 228.

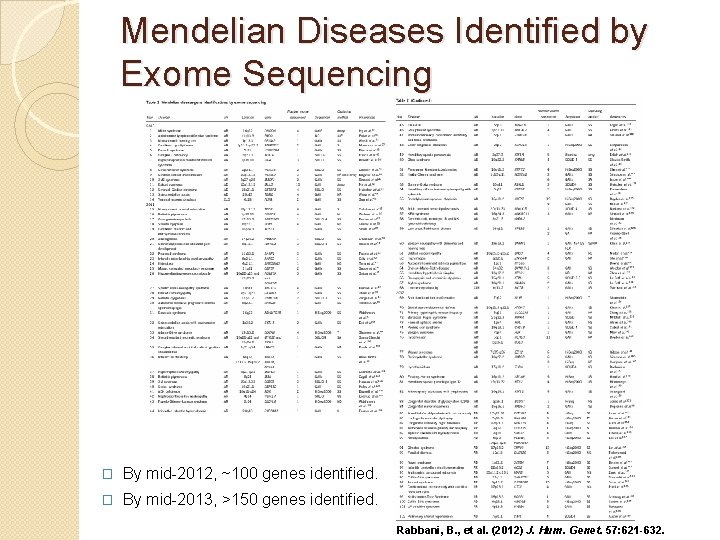
Mendelian Diseases Identified by Exome Sequencing � By mid-2012, ~100 genes identified. � By mid-2013, >150 genes identified. Rabbani, B. , et al. (2012) J. Hum. Genet. 57: 621 -632.

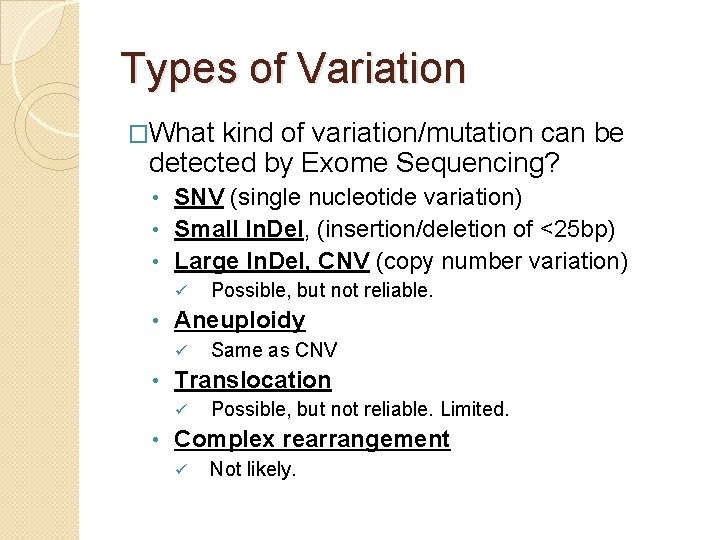
Types of Variation �What kind of variation/mutation can be detected by Exome Sequencing? SNV (single nucleotide variation) • Small In. Del, (insertion/deletion of <25 bp) • Large In. Del, CNV (copy number variation) • ü • Aneuploidy ü • Same as CNV Translocation ü • Possible, but not reliable. Limited. Complex rearrangement ü Not likely.

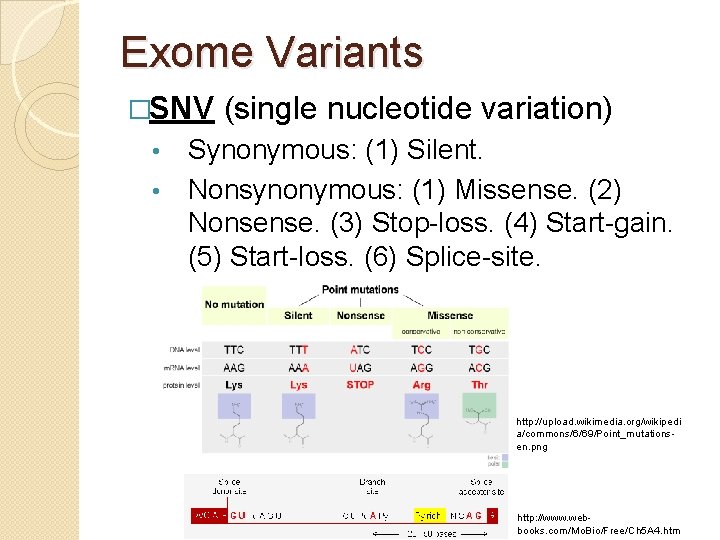
Exome Variants �SNV (single nucleotide variation) Synonymous: (1) Silent. • Nonsynonymous: (1) Missense. (2) Nonsense. (3) Stop-loss. (4) Start-gain. (5) Start-loss. (6) Splice-site. • http: //upload. wikimedia. org/wikipedi a/commons/6/69/Point_mutationsen. png http: //www. webbooks. com/Mo. Bio/Free/Ch 5 A 4. htm

Exome Variants �Small In. Del (insertion/deletion <25 bp) Frameshift • In-frame • NHGRI Digital Media Database (DMD), http: //www. genome. gov/dmd/

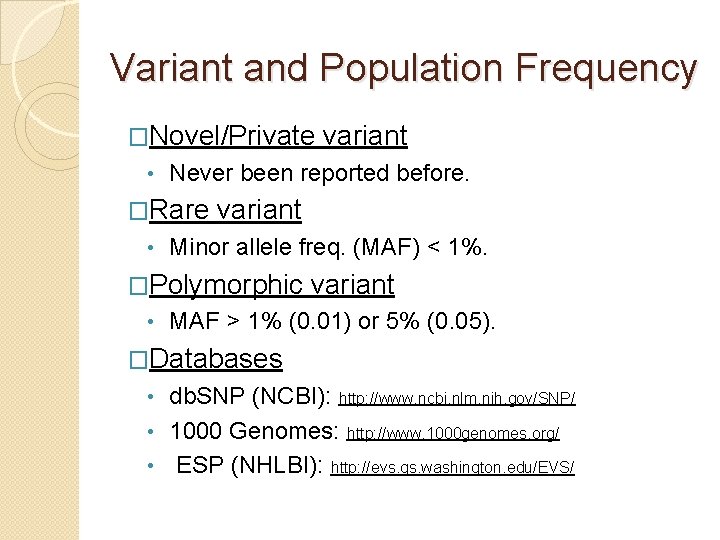
Variant and Population Frequency �Novel/Private • Never been reported before. �Rare • variant Minor allele freq. (MAF) < 1%. �Polymorphic • variant MAF > 1% (0. 01) or 5% (0. 05). �Databases db. SNP (NCBI): http: //www. ncbi. nlm. nih. gov/SNP/ • 1000 Genomes: http: //www. 1000 genomes. org/ • ESP (NHLBI): http: //evs. gs. washington. edu/EVS/ •

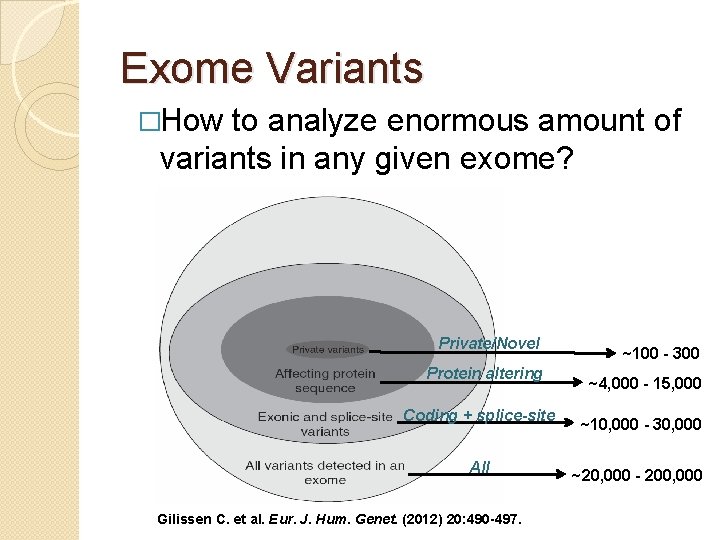
Exome Variants �How to analyze enormous amount of variants in any given exome? Private/Novel Protein altering Coding + splice-site All Gilissen C. et al. Eur. J. Hum. Genet. (2012) 20: 490 -497. ~100 - 300 ~4, 000 - 15, 000 ~10, 000 - 30, 000 ~20, 000 - 200, 000

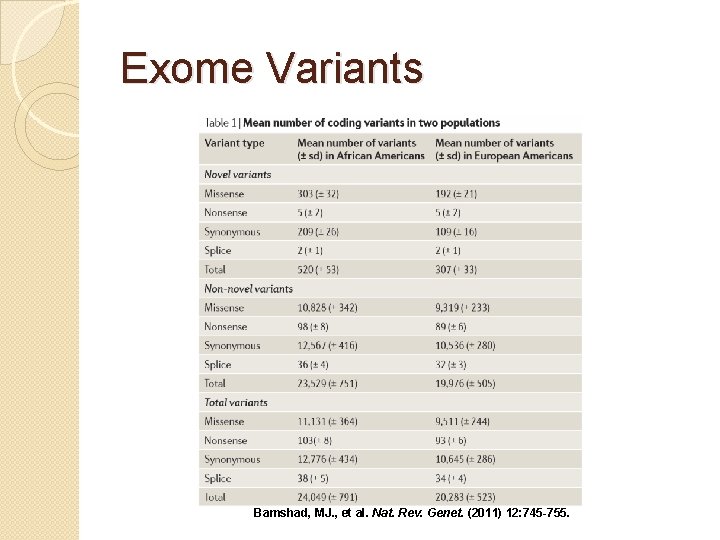
Exome Variants Bamshad, MJ. , et al. Nat. Rev. Genet. (2011) 12: 745 -755.

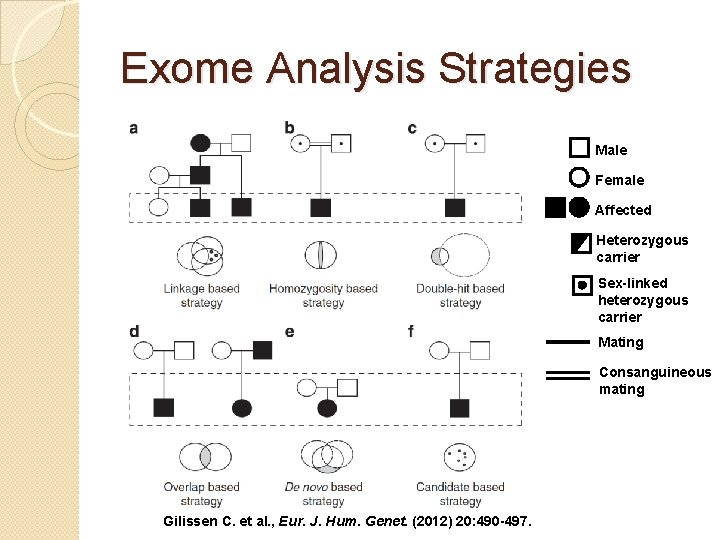
Exome Analysis Strategies Male Female Affected Heterozygous carrier Sex-linked heterozygous carrier Mating Consanguineous mating Gilissen C. et al. , Eur. J. Hum. Genet. (2012) 20: 490 -497.

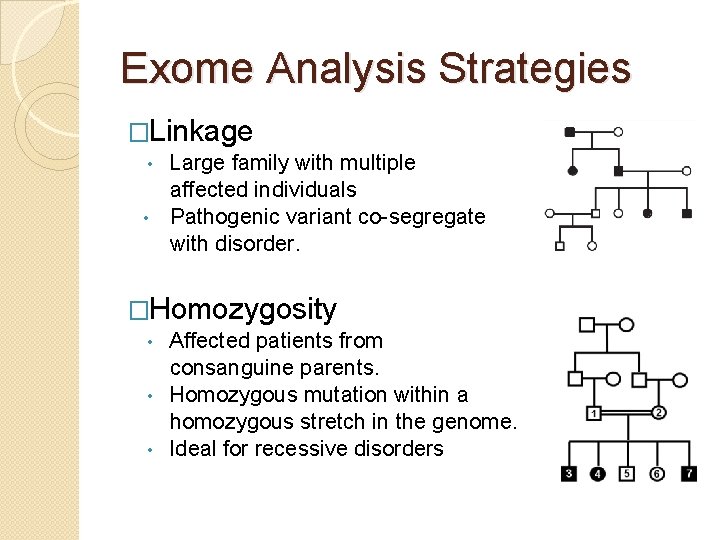
Exome Analysis Strategies �Linkage Large family with multiple affected individuals • Pathogenic variant co-segregate with disorder. • �Homozygosity Affected patients from consanguine parents. • Homozygous mutation within a homozygous stretch in the genome. • Ideal for recessive disorders •

Exome Analysis Strategies �Candidate genes Biased approach • Require current biological knowledge • Good for screening or clinical diagnosis of known disorders. • �Overlap Require multiple unrelated individuals with identical disorders. • Monogenic disorders •

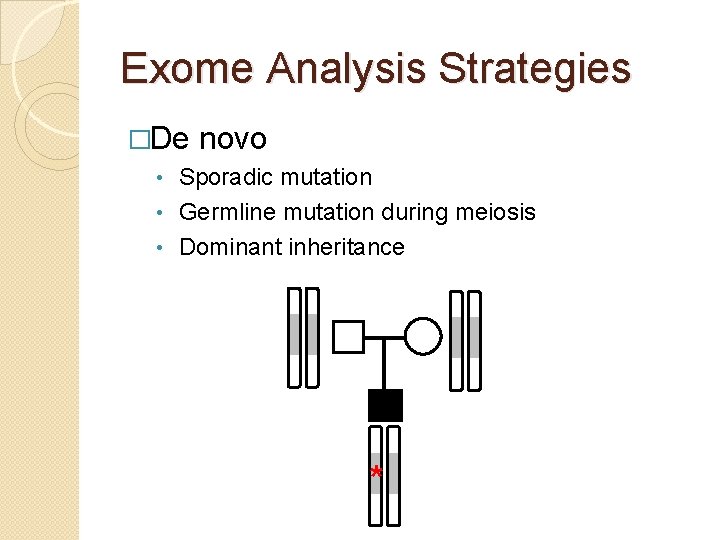
Exome Analysis Strategies �De novo Sporadic mutation • Germline mutation during meiosis • Dominant inheritance • *

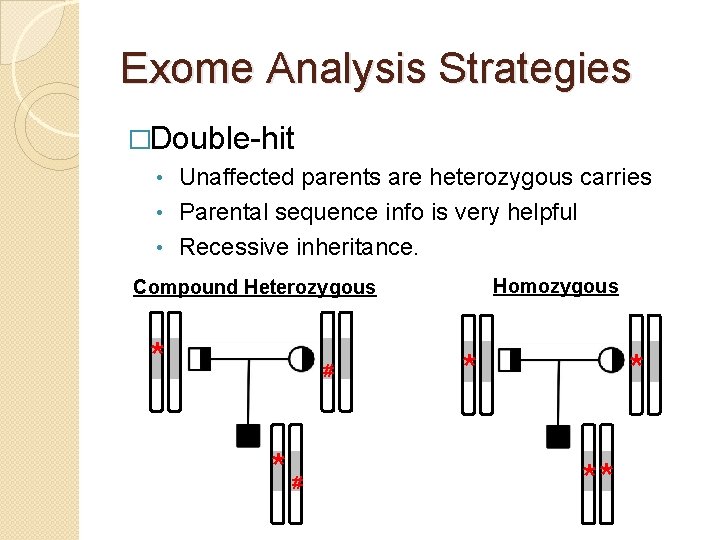
Exome Analysis Strategies �Double-hit Unaffected parents are heterozygous carries • Parental sequence info is very helpful • Recessive inheritance. • Homozygous Compound Heterozygous * # *# * * **

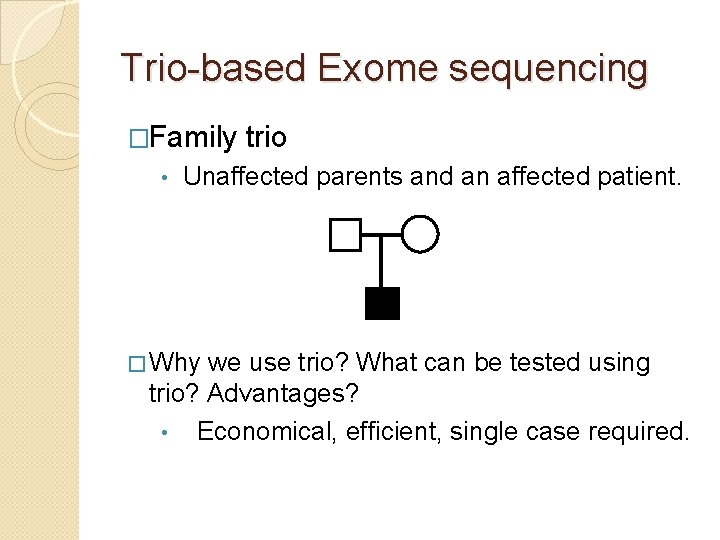
Trio-based Exome sequencing �Family • trio Unaffected parents and an affected patient. � Why we use trio? What can be tested using trio? Advantages? • Economical, efficient, single case required.

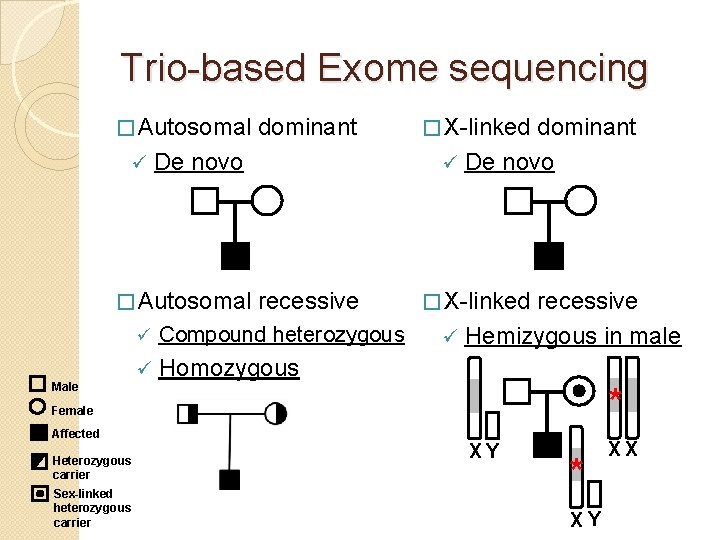
Trio-based Exome sequencing � Autosomal ü � X-linked recessive � X-linked De novo � Autosomal Male dominant ü Compound heterozygous ü Homozygous dominant ü De novo recessive ü Hemizygous in male * Female Affected Heterozygous carrier Sex-linked heterozygous carrier XY * XY XX

Trio-based Exome sequencing �Candidate Genes/Variants Protein altering variants • Rare or novel variants • Variants that fit each inheritance model • Dominant Recessive Rare Variant Novel Variant De novo 0~1 Compound Heterozygous 0 ~ 20 0~3 Homozygous 0 ~ 20 0~3 X-linked 0 ~ 10 0~5

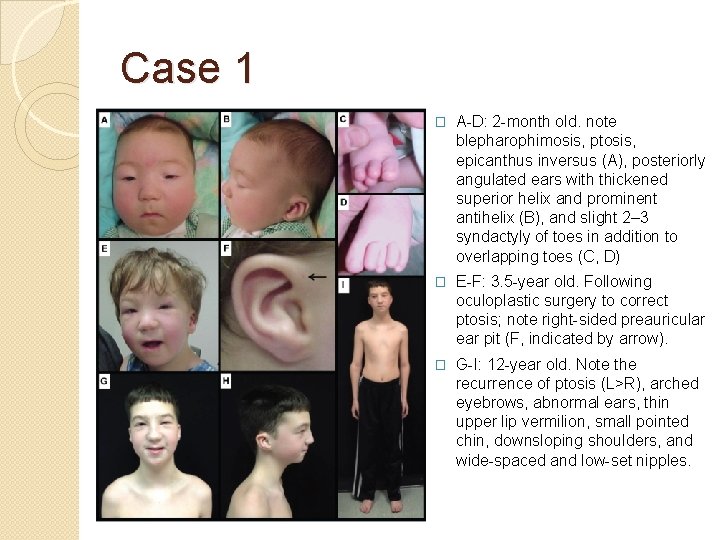
Case 1 �Clinical information The patient was a 7 -month-old boy when first evaluated. He was diagnosed with BPES by a pediatric ophthalmologist. In addition to blepharophimosis, ptosis, and epicanthus inversus normally associated with BPES, he had cryptorchidism, right hydrocele, wide -spaced nipples, and slight 2– 3 syndactyly of toes. Clinical testing demonstrated a normal karyotype (46, XY), and normal FISH studies for 22 q 11. 2 deletion, Cri-du-Chat (5 p deletion) syndrome. Thyroid function was normal. Further, normal 7 -dehydrocholesterol level was used to rule out Smith–Lemli–Opitz syndrome. Sanger sequencing and highresolution CNV analysis with Affymetrix SNP 500 K arrays did not identify a FOXL 2 mutation.

Case 1 � A-D: 2 -month old. note blepharophimosis, ptosis, epicanthus inversus (A), posteriorly angulated ears with thickened superior helix and prominent antihelix (B), and slight 2– 3 syndactyly of toes in addition to overlapping toes (C, D) � E-F: 3. 5 -year old. Following oculoplastic surgery to correct ptosis; note right-sided preauricular ear pit (F, indicated by arrow). � G-I: 12 -year old. Note the recurrence of ptosis (L>R), arched eyebrows, abnormal ears, thin upper lip vermilion, small pointed chin, downsloping shoulders, and wide-spaced and low-set nipples.

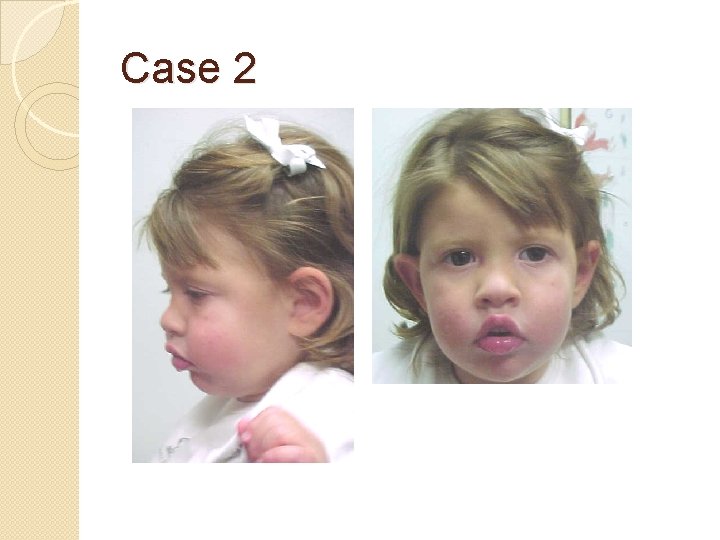
Case 2 �Clinical information The proband is a nine year old girl who presented with microcephaly, unilateral retinal coloboma, bilateral optic nerve hypoplasia, nystagmus, seizures, gastroesophageal reflux, and developmental delay including not yet saying specific words (at 29 months old). On exam, she has microcephaly with a normal height, a down-turned upper lip, and fingertip pads. A karyotype and CGH analysis have been normal. Kabuki (KMT 2 D and KDM 6 A) and Angelman (UBE 3 A and MECP 2) syndromes were suspected in this patient.

Case 2

Case 3 �Clinical information Case 3 was the result of a non-consanguineous union and he presented to care at four months of age with a seizure disorder, hypotonia and developmental delay. The patient underwent a left parietal craniotomy and partial resection of the frontal cortex without complete resolution of the seizure disorder. Initial laboratory studies included an elevated homocysteine and methylmalonic acid and a normal vitamin B 12 level. Complementation analysis of the patient’s cell line placed the patient into the cbl. C class. Sequencing and deletion/duplication analysis (microarray) the MMACHC gene was negative in both skin fibroblasts and peripheral blood.

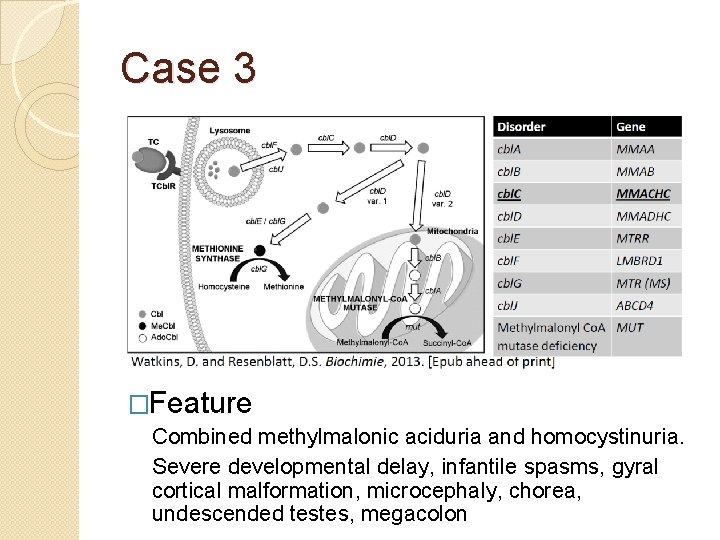
Case 3 �Feature Combined methylmalonic aciduria and homocystinuria. Severe developmental delay, infantile spasms, gyral cortical malformation, microcephaly, chorea, undescended testes, megacolon

Case 3 �Monster Max http: //www. maxwatson. org/ �Patient's older sister as a summer student in Shaikh Lab

Data for Case Study � 3 • • trios A total of 3 families/cases. Each family/case includes unaffected parents and an affected patient. �VCF • • files Familial variants calls in VCF format, mapped to human GRCh 37/hg 19. 2 x 90 bp paired-end reads, with ~50 X coverage �“Mini” • • Exome 100 genes with/without known disorder association. Validated causative genes, plus randomly selected genes.

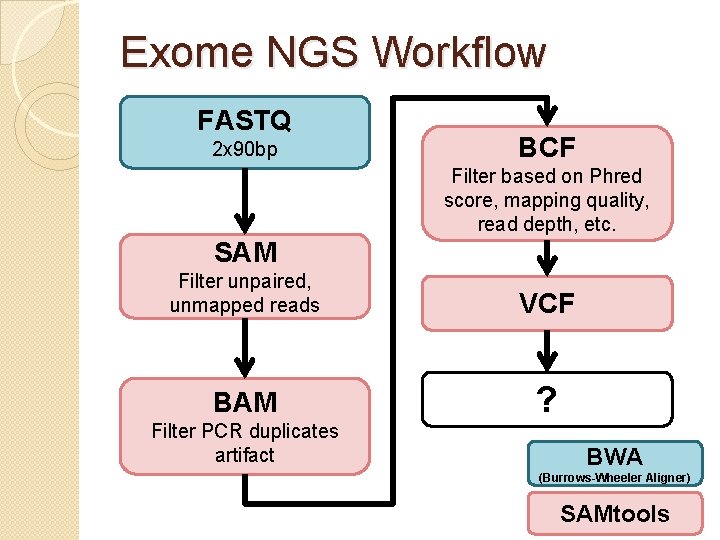
Exome NGS Workflow FASTQ 2 x 90 bp BCF Filter based on Phred score, mapping quality, read depth, etc. SAM Filter unpaired, unmapped reads VCF BAM ? Filter PCR duplicates artifact BWA (Burrows-Wheeler Aligner) SAMtools

VCF Format �VCF (Variant Call Format) http: //www. 1000 genomes. org/wiki/Analysis/Variant%20 Call%20 Format/vcf-variant-call-format-version-41 ## Meta-information lines FILTER, INFO, FORMAT # Header line

VCF Format � INFO � AA : ancestral allele � AC : allele count in genotypes, for each ALT allele, in the same order as listed � AF : allele frequency for each ALT allele in the same order as listed: use this when estimated from primary data, not called genotypes � AN : total number of alleles in called genotypes � BQ : RMS base quality at this position � CIGAR : cigar string describing how to align an alternate allele to the reference allele � DB : db. SNP membership � DP : combined depth across samples, e. g. DP=154 � END : end position of the variant described in this record (for use with symbolic alleles) � H 2 : membership in hapmap 2 � H 3 : membership in hapmap 3 � MQ : RMS mapping quality, e. g. MQ=52 � MQ 0 : Number of MAPQ == 0 reads covering this record � NS : Number of samples with data � SB : strand bias at this position � SOMATIC : indicates that the record is a somatic mutation, for cancer genomics � VALIDATED : validated by follow-up experiment � 1000 G : membership in 1000 Genomes

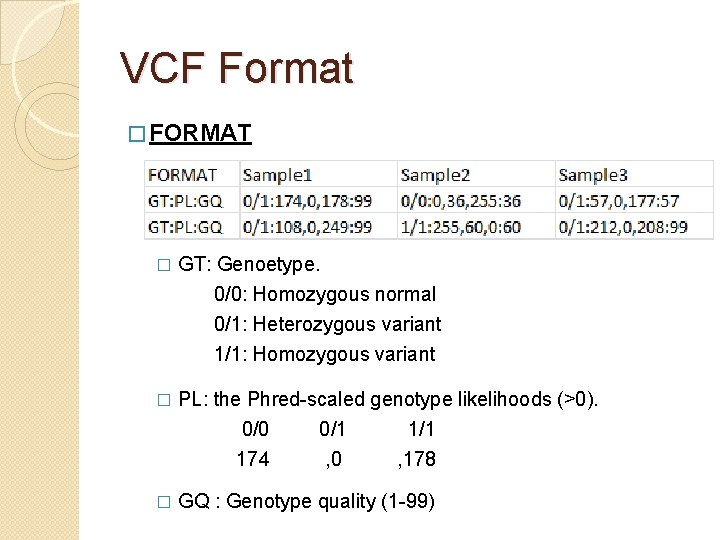
VCF Format � FORMAT � GT: Genoetype. 0/0: Homozygous normal 0/1: Heterozygous variant 1/1: Homozygous variant � PL: the Phred-scaled genotype likelihoods (>0). 0/0 0/1 174 , 0 , 178 � GQ : Genotype quality (1 -99)

Question ?
 Exome sequencing project
Exome sequencing project Family resemblance test
Family resemblance test Exome
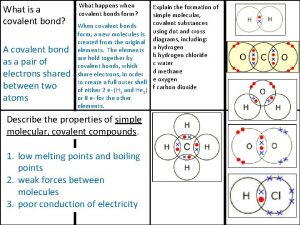
Exome Covalent bond
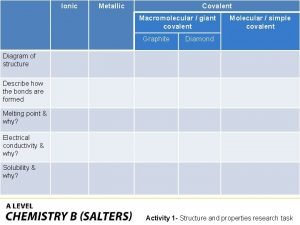
Covalent bond Giant molecular structure vs simple molecular structure
Giant molecular structure vs simple molecular structure Giant molecular structure vs simple molecular structure
Giant molecular structure vs simple molecular structure Loopback adapter
Loopback adapter Husqvarna autotune diagnostic tool software download
Husqvarna autotune diagnostic tool software download Remote desktop services diagnostic tool
Remote desktop services diagnostic tool Chapter 10 section 2 mendelian genetics answer key
Chapter 10 section 2 mendelian genetics answer key Chapter 7 extending mendelian genetics vocabulary practice
Chapter 7 extending mendelian genetics vocabulary practice Inheritance
Inheritance Mendelian ratios
Mendelian ratios Probability laws govern mendelian inheritance
Probability laws govern mendelian inheritance Exceptions to mendelian genetics
Exceptions to mendelian genetics Non mendelian law
Non mendelian law 11-3 exploring mendelian genetics
11-3 exploring mendelian genetics Multiple alleles
Multiple alleles Allelic and non allelic interaction
Allelic and non allelic interaction Section 11-4 meiosis answer key
Section 11-4 meiosis answer key What are mendel's three laws of inheritance
What are mendel's three laws of inheritance Mendelian traits
Mendelian traits Mendel was a
Mendel was a Dominant or recessive
Dominant or recessive Chromosomes and alleles
Chromosomes and alleles Non mendelian inheritance
Non mendelian inheritance Mendelian genetics definition
Mendelian genetics definition Section 1 chromosomes and phenotype study guide a
Section 1 chromosomes and phenotype study guide a Non mendelian law
Non mendelian law Chapter 7 extending mendelian genetics study guide answers
Chapter 7 extending mendelian genetics study guide answers X-linked recessive punnett square
X-linked recessive punnett square 9:3:3:1 ratio
9:3:3:1 ratio What is heterozygous type b blood
What is heterozygous type b blood Chapter 10 sexual reproduction and genetics
Chapter 10 sexual reproduction and genetics Pprr x pprr punnett square
Pprr x pprr punnett square 11.3 exploring mendelian genetics
11.3 exploring mendelian genetics Mendelian
Mendelian Mendelian principles
Mendelian principles Codominance example
Codominance example