Dr Heather Miller Coyle People v Jaquan Collins














- Slides: 25

Dr. Heather Miller Coyle People v. Jaquan Collins People v. Andrew Peaks Nov. 8, 2013

Validation p Purpose of validation n To ensure a method is reliable and robust and reproducible for use in casework n Requisite for new methodologies n Reliable method defined as one where results are accurate and reflect the sample being tested n Reproducible method defined as the same or very similar results obtained each time tested

Validation p p p Purpose of validation Developmental validation – method development Internal validation – a method validated in-house in a lab means it is generally accepted in the lab but not necessarily accepted by the scientific community as reliable n If accepted by scientific community, general use by all laboratories would follow n Low copy number (LCN) is problematic and not generally used in criminal casework across the United States

Scope of Peer Review 1. p Peer review is open scientific evaluation by appropriate scientific community as a step toward general acceptance or rejection of method Dr. Caragine’s testimony: n n p p Question: And then when an article is published in the peer review journal, what does that mean? Answer: It means that your results have been accepted by the sci-by the your peers, by the scientific community. Publication in a peer reviewed journal does not equal general acceptance Publication in peer reviewed journal does not equal exhaustive peer review since all of the data was not submitted to journal (summary)

Auditing Is Not Peer Review Auditing is a quality assurance/quality control process p It is aimed at ensuring that a laboratory follows protocols and the DNA Advisory Board standards and is not primarily aimed at ensuring that the underlying scientific methodology is reliable p

What is Low Copy Number Testing? p p Methodology employing increased sensitivity techniques on low template samples (<100 pg) Generally accepted forensic DNA tests employ 28 cycles of a PCR method LCN uses 31 cycles of PCR (cycle enhancement) and different interpretation criteria for samples and controls (not the same method) Samples and controls should be reproducible from PCR amplification to PCR amplification but are not due to low template concentrations of DNA (<100 pg) (stochastic effects)

Characteristics of Low Copy Number DNA Testing: Stochastic Effects Stochastic effects: Ø Ø Ø Increased heterozygote peak imbalance “Drop-out” (extreme form of PHI) Increased baseline Increased stutter (artifactual PCR peaks) Increased “drop-in” or contamination “Drop-in” and “drop-out” confounds the ability to determine with accuracy the true number of contributors (source attribution error in LCN)

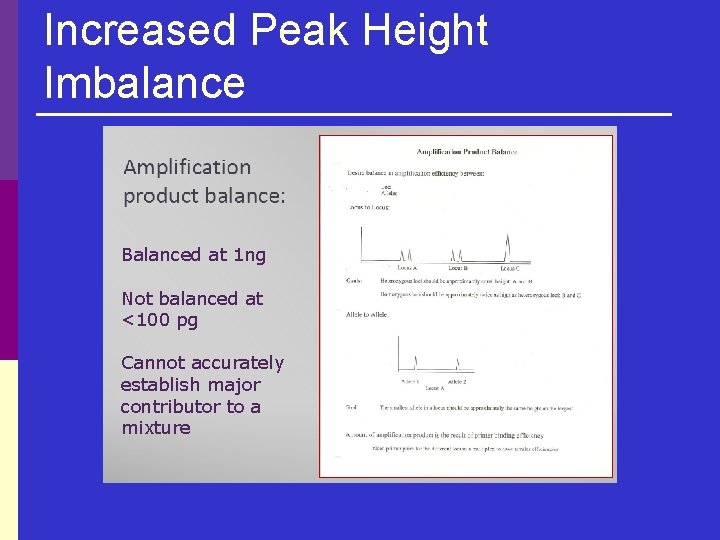
Increased Peak Height Imbalance Balanced at 1 ng Not balanced at <100 pg Cannot accurately establish major contributor to a mixture

Increased Peak Height Imbalance p “Drop out” is an extreme example n p Drop out rate is especially variable in the stochastic range of under 100 pg, particularly below 50 pg Affects interpretation of results how n n False homozygote Difficulty in deconvoluting mixtures with accuracy May incorrectly conclude two peaks are “sisters”, masking Error rates or false inclusion rate is high


Characteristics of Low Copy Number DNA Testing: “Drop-In” p p p “Drop-In” is contamination OCME defines as “drop-in” as observation of a single DNA peak once in triplicate amplifications; “contamination” is if DNA peak is observed more than once Problematic to distinguish between authentic DNA representative of sample and contamination from an outside source Clean facilities and UV treatment of tubes still yields 8 -11% extraneous DNA in negative controls Extremely confounding when interpreting DNA results

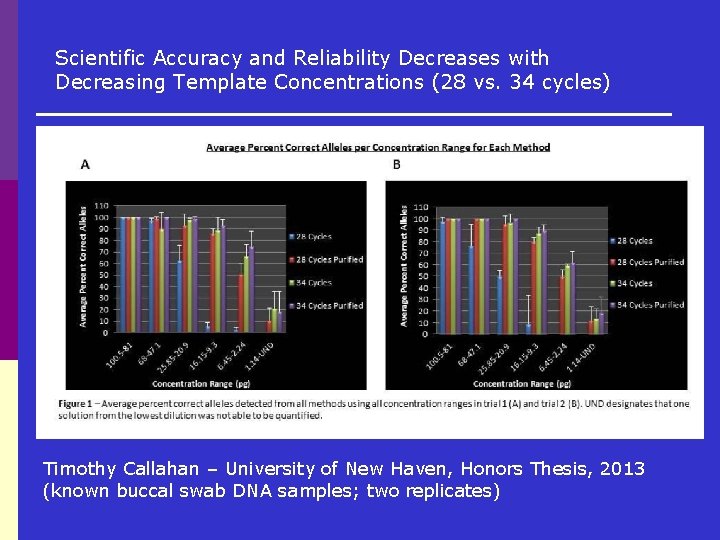
Scientific Accuracy and Reliability Decreases with Decreasing Template Concentrations (28 vs. 34 cycles) Timothy Callahan – University of New Haven, Honors Thesis, 2013 (known buccal swab DNA samples; two replicates)

Complex Mixtures Add Another Layer of Uncertainty p p p Complex mixture interpretation is difficult enough at 28 cycle, high template DNA testing Controversy in scientific community about how to interpret and report complex DNA mixtures Low level LCN testing is that much worse Stochastic effects make it difficult to establish number of true contributors and major donors Coincidental matching makes it difficult to identify which DNA fragments go to which individual

Consequence of Underestimating # of Contributors and/or Deconvoluting Mixtures Underestimating number of contributors increases the source attribution error (force fit of data) p Source attribution error = deducing profiles (major donor) from a mixture is not valid in stochastic range p Both result in scientific unreliability or scientific inaccuracy of a result p

Negative Control Contamination p p Purpose of negative controls is to establish whether extraneous DNA is present that was not originally in the sample OCME-NYC allows up to 9 spurious alleles in the negative controls before contamination is at an unacceptable level LCN validation data shows different injections, resulting in different alleles detected in negative controls Not generally accepted to have contamination in negative controls

OCME Performed Ten Touch Studies p Six clean studies (used various items: hands, pens, CD case, lunch box, etc. ) n p Two studies were mixed (clean and dirty) n n p One study was performed using three-person touched items; the second study used four-person touched items. These studies had nine clean items and nine dirty items. One study used dirty items n p These six studies were performed using two-, three-, and four-person touched items. These were two person touched items. One study was performed on degraded items

Clean Touch Studies p Examined 85 clean touch samples when calculating alleles that were unaccounted for by the known contributors. n n ID 28 = 41 samples ID 31 = 44 samples Summaries 19, 20, and 21: designed to replicate case work protocols followed in cleaning items before items were touched. p OCME cleaning protocols unable to eliminate contamination. p

Analysis p Determined the number of unaccounted alleles in n n 28 cycles 31 cycles Total # of unaccounted alleles/(# samples x # of replicates x # loci) = % of detected alleles that did not come from a known contributor

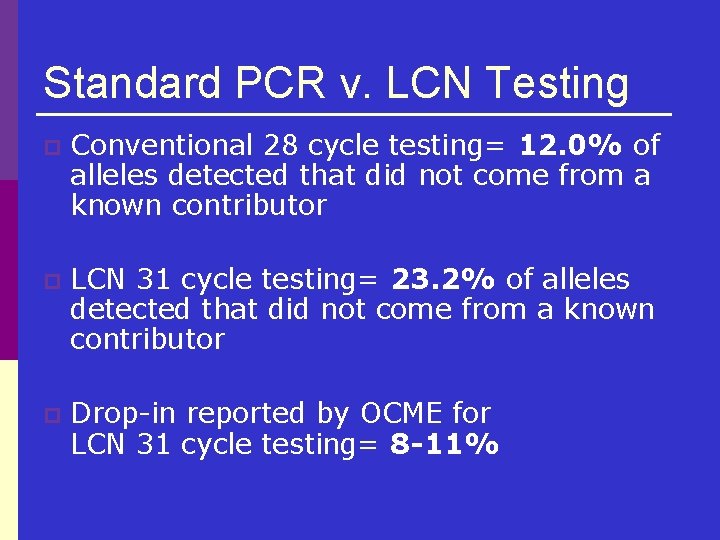
Standard PCR v. LCN Testing p Conventional 28 cycle testing= 12. 0% of alleles detected that did not come from a known contributor p LCN 31 cycle testing= 23. 2% of alleles detected that did not come from a known contributor p Drop-in reported by OCME for LCN 31 cycle testing= 8 -11%

Contamination Percentage for Total Allele Count p Calculated the percentage of contaminant alleles to the total allele count for each mixture.

Studies 2 A and 2 B (estimated % contaminant alleles) p p p ID 31 Hand 2 Hand 4 Hand 5 Hand 18 2 P Pen. D 2 P Pen. E 2 P Pen. G 2 P Pen. H 2 P Pen. J 2 P Mouse Ave. %: ID 28 7. 6% 35. 0% 26. 0% 30. 0% 8. 9% 11. 9% 0. 0% 7. 6% 16. 5% 21. 4% 16. 0% Hand 3 0% Hand 9 2. 4% Hand 10 2. 7% Hand 11 4. 4% Hand 14 16. 8% Hand 15 3. 7% Hand 16 0% 2 P Stapler 1. 7% 2 P CD case 0% 2 P plastic cover 0% Ave. %: 2. 3%

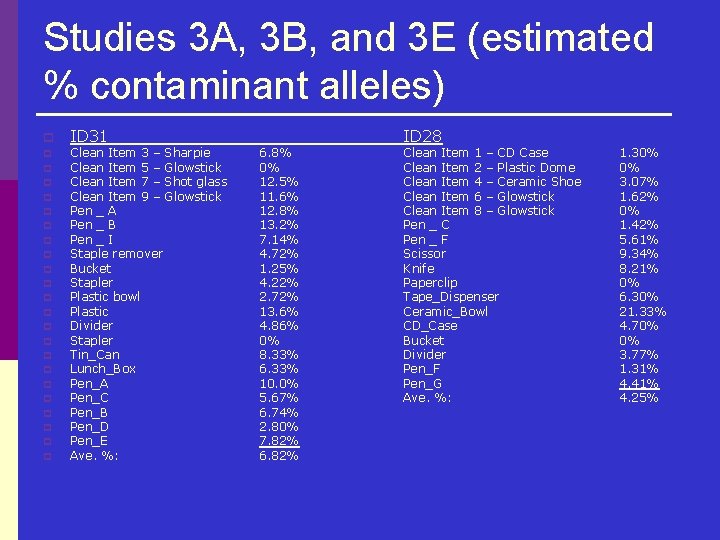
Studies 3 A, 3 B, and 3 E (estimated % contaminant alleles) p p p p p p ID 31 Clean Item 3 – Sharpie Clean Item 5 – Glowstick Clean Item 7 – Shot glass Clean Item 9 – Glowstick Pen _ A Pen _ B Pen _ I Staple remover Bucket Stapler Plastic bowl Plastic Divider Stapler Tin_Can Lunch_Box Pen_A Pen_C Pen_B Pen_D Pen_E Ave. %: 6. 8% 0% 12. 5% 11. 6% 12. 8% 13. 2% 7. 14% 4. 72% 1. 25% 4. 22% 2. 72% 13. 6% 4. 86% 0% 8. 33% 6. 33% 10. 0% 5. 67% 6. 74% 2. 80% 7. 82% 6. 82% ID 28 Clean Item 1 – CD Case Clean Item 2 – Plastic Dome Clean Item 4 – Ceramic Shoe Clean Item 6 – Glowstick Clean Item 8 – Glowstick Pen _ C Pen _ F Scissor Knife Paperclip Tape_Dispenser Ceramic_Bowl CD_Case Bucket Divider Pen_F Pen_G Ave. %: 1. 30% 0% 3. 07% 1. 62% 0% 1. 42% 5. 61% 9. 34% 8. 21% 0% 6. 30% 21. 33% 4. 70% 0% 3. 77% 1. 31% 4. 41% 4. 25%

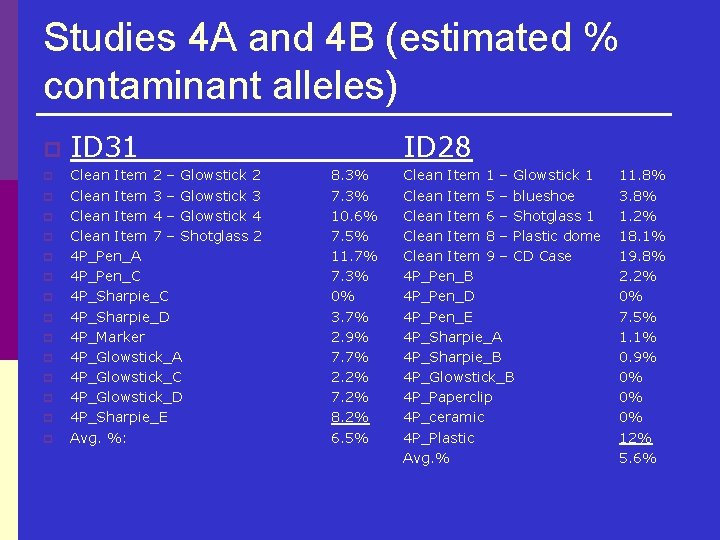
Studies 4 A and 4 B (estimated % contaminant alleles) p p p p ID 31 Clean Item 2 – Glowstick 2 Clean Item 3 – Glowstick 3 Clean Item 4 – Glowstick 4 Clean Item 7 – Shotglass 2 4 P_Pen_A 4 P_Pen_C 4 P_Sharpie_D 4 P_Marker 4 P_Glowstick_A 4 P_Glowstick_C 4 P_Glowstick_D 4 P_Sharpie_E Avg. %: ID 28 8. 3% 7. 3% 10. 6% 7. 5% 11. 7% 7. 3% 0% 3. 7% 2. 9% 7. 7% 2. 2% 7. 2% 8. 2% 6. 5% Clean Item 1 – Glowstick 1 Clean Item 5 – blueshoe Clean Item 6 – Shotglass 1 Clean Item 8 – Plastic dome Clean Item 9 – CD Case 4 P_Pen_B 4 P_Pen_D 4 P_Pen_E 4 P_Sharpie_A 4 P_Sharpie_B 4 P_Glowstick_B 4 P_Paperclip 4 P_ceramic 4 P_Plastic Avg. % 11. 8% 3. 8% 1. 2% 18. 1% 19. 8% 2. 2% 0% 7. 5% 1. 1% 0. 9% 0% 0% 0% 12% 5. 6%

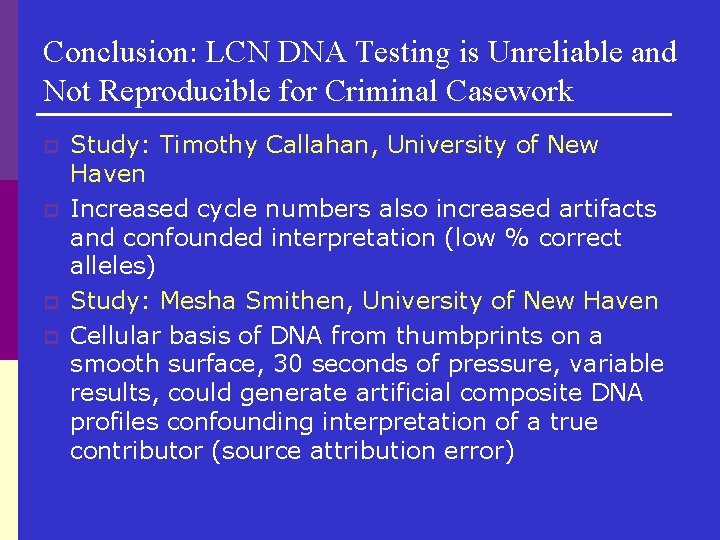
Conclusion: LCN DNA Testing is Unreliable and Not Reproducible for Criminal Casework p p Study: Timothy Callahan, University of New Haven Increased cycle numbers also increased artifacts and confounded interpretation (low % correct alleles) Study: Mesha Smithen, University of New Haven Cellular basis of DNA from thumbprints on a smooth surface, 30 seconds of pressure, variable results, could generate artificial composite DNA profiles confounding interpretation of a true contributor (source attribution error)

Lack of Scientific Accuracy and General Acceptance of LCN Use in Forensic Community p p p Other forensic science laboratories do not utilize this LCN process in United States “Drop-out” rates make it difficult to establish number of contributors with accuracy Contamination rates are high (more than 8 -11%) so DNA sample does not accurately reflect contributor Kits are not optimized to <100 pg so stochastic effects and PCR artifacts are high and confounds interpretation Statistics are challenging with results being inconclusive or with FST software, a high false inclusion rates due to coincidental matching of DNA
 Jaquan collins
Jaquan collins Dr heather collins
Dr heather collins Heather james ice miller
Heather james ice miller Medical plaza miller 131 miller street
Medical plaza miller 131 miller street Miller index
Miller index Daniel miller arthur miller
Daniel miller arthur miller Coyle
Coyle Dr christine coyle
Dr christine coyle Caitlin coyle
Caitlin coyle Coyle health and wellbeing
Coyle health and wellbeing Metodo coyle
Metodo coyle Do coyle clil
Do coyle clil Aaron coyle
Aaron coyle Coyle electric
Coyle electric Coyle method
Coyle method Coyle bernard
Coyle bernard Do coyle
Do coyle Coyle method
Coyle method People just people
People just people Who are the people involved in media
Who are the people involved in media Where is the love lyrics 2016
Where is the love lyrics 2016 Recovery community
Recovery community Heather sweeney designs
Heather sweeney designs Heather wakelee
Heather wakelee Heather blackwood
Heather blackwood Heather gamble
Heather gamble