DEMRI Dynamic Contrast Enhanced MRI Kinetic modeling ARN


- Slides: 12

DEMRI (Dynamic Contrast Enhanced MRI) Kinetic modeling



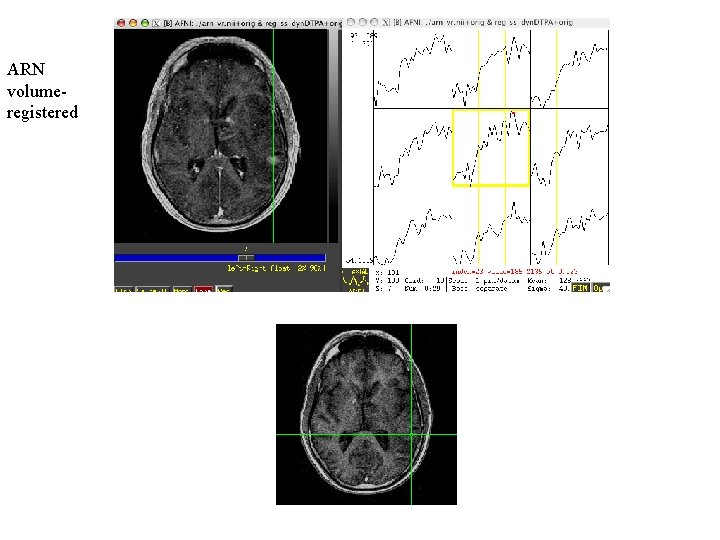
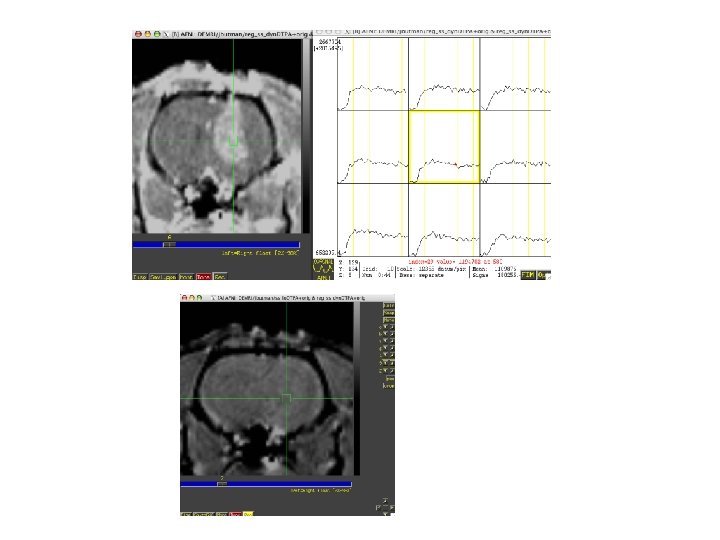
ARN volumeregistered

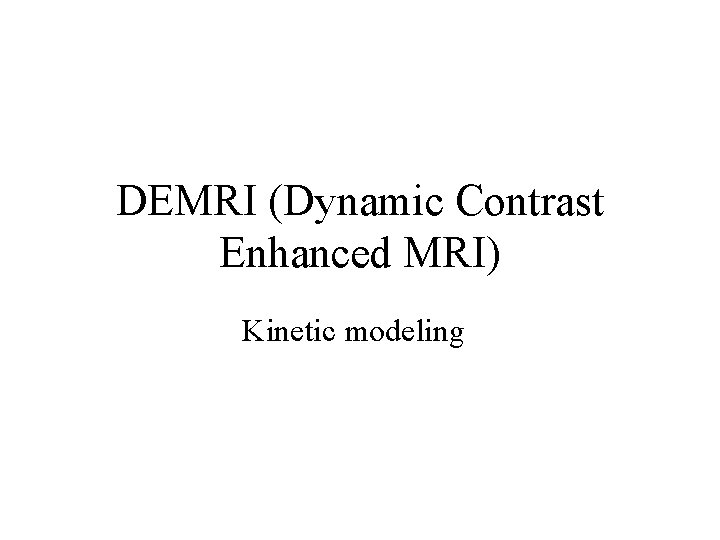
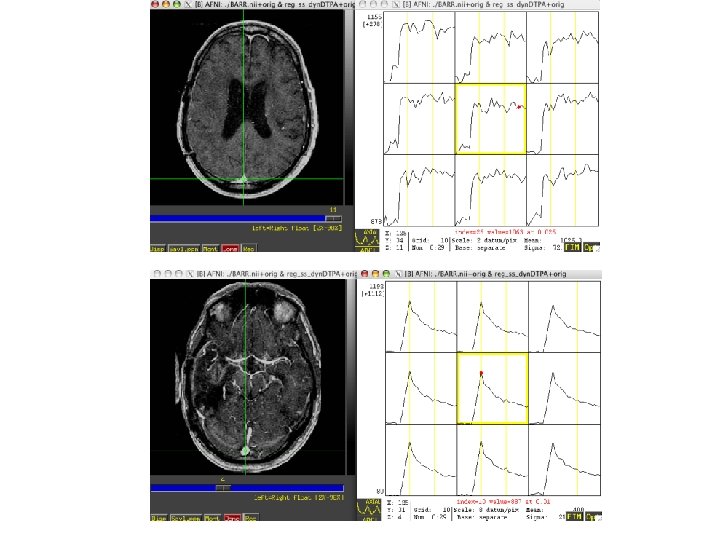
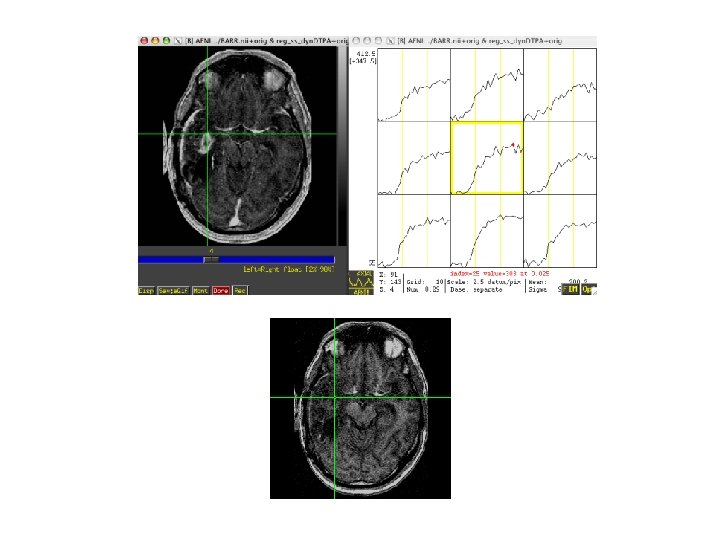
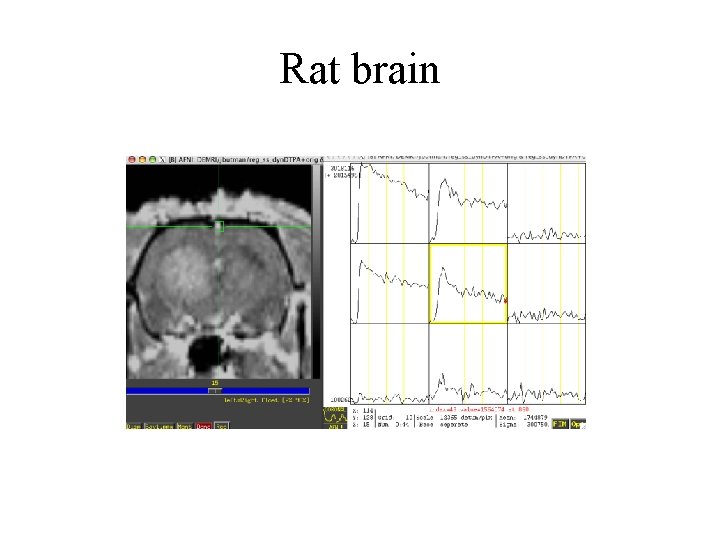
Rat brain


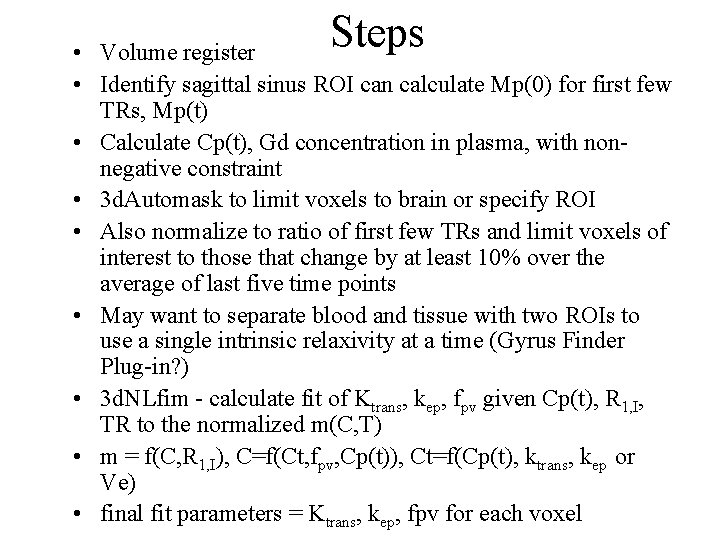
Steps • Volume register • Identify sagittal sinus ROI can calculate Mp(0) for first few TRs, Mp(t) • Calculate Cp(t), Gd concentration in plasma, with nonnegative constraint • 3 d. Automask to limit voxels to brain or specify ROI • Also normalize to ratio of first few TRs and limit voxels of interest to those that change by at least 10% over the average of last five time points • May want to separate blood and tissue with two ROIs to use a single intrinsic relaxivity at a time (Gyrus Finder Plug-in? ) • 3 d. NLfim - calculate fit of Ktrans, kep, fpv given Cp(t), R 1, I, TR to the normalized m(C, T) • m = f(C, R 1, I), C=f(Ct, fpv, Cp(t)), Ct=f(Cp(t), ktrans, kep or Ve) • final fit parameters = Ktrans, kep, fpv for each voxel

----- HISTORY ----================== === History of inputs to 3 dcalc === Input a: 3 dvolreg -prefix barr_vr. nii BARR. nii === Input b: 3 dvolreg -prefix barr_vr. nii BARR. nii 3 d. Tstat -prefix barr_mean 7. nii 'barr_vr. nii[0. . 6]' === Input c: 3 dvolreg -prefix barr_vr. nii BARR. nii 3 d. Tstat -prefix barr_mean 7. nii 'barr_vr. nii[0. . 6]' 3 d. Automask -prefix barr_mask barr_mean 7. nii ================== 3 dcalc -a barr_vr. nii -b barr_mean 7. nii -c barr_mask+orig -prefix barr_norm 7 m. nii -datum float -expr 'a*c/b' 3 d. NLfim -input barr_norm 7 m. nii -signal demri_3 -ignore 0 -noise Zero -sconstr 0 0 0. 05 -sconstr 1 0 0. 05 -nabs -mask barr_mask+orig -nrand 10000 -nbest 10 -voxel_count -sfit barr_sfit 6 -jobs 2 -progress 10000 -bucket 0 nl 6. out Output prefix: nl 6. out

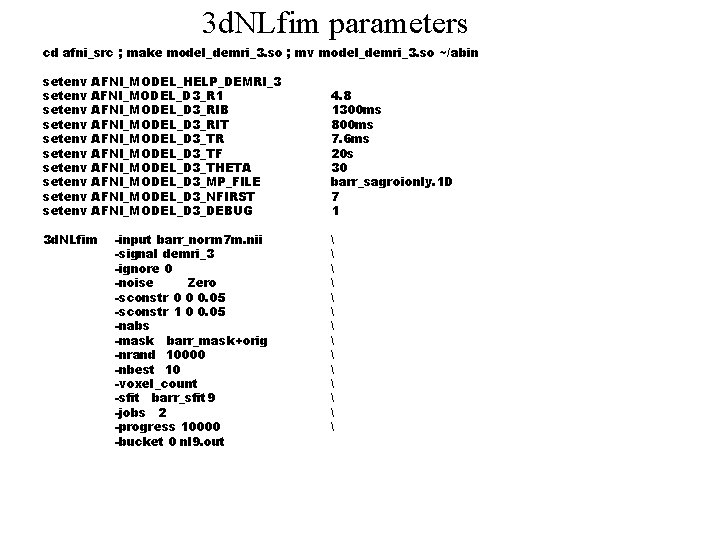
3 d. NLfim parameters cd afni_src ; make model_demri_3. so ; mv model_demri_3. so ~/abin setenv setenv setenv AFNI_MODEL_HELP_DEMRI_3 AFNI_MODEL_D 3_R 1 AFNI_MODEL_D 3_RIB AFNI_MODEL_D 3_RIT AFNI_MODEL_D 3_TR AFNI_MODEL_D 3_TF AFNI_MODEL_D 3_THETA AFNI_MODEL_D 3_MP_FILE AFNI_MODEL_D 3_NFIRST AFNI_MODEL_D 3_DEBUG 3 d. NLfim -input barr_norm 7 m. nii -signal demri_3 -ignore 0 -noise Zero -sconstr 0 0 0. 05 -sconstr 1 0 0. 05 -nabs -mask barr_mask+orig -nrand 10000 -nbest 10 -voxel_count -sfit barr_sfit 9 -jobs 2 -progress 10000 -bucket 0 nl 9. out 4. 8 1300 ms 800 ms 7. 6 ms 20 s 30 barr_sagroionly. 1 D 7 1

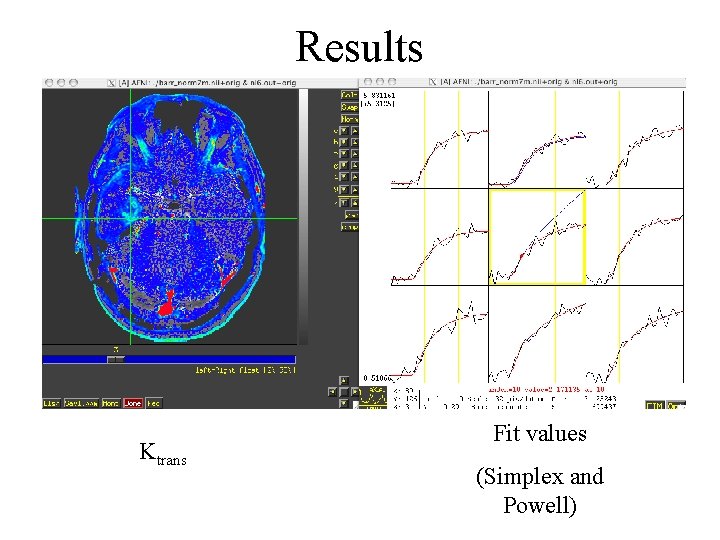
Results Ktrans Fit values (Simplex and Powell)

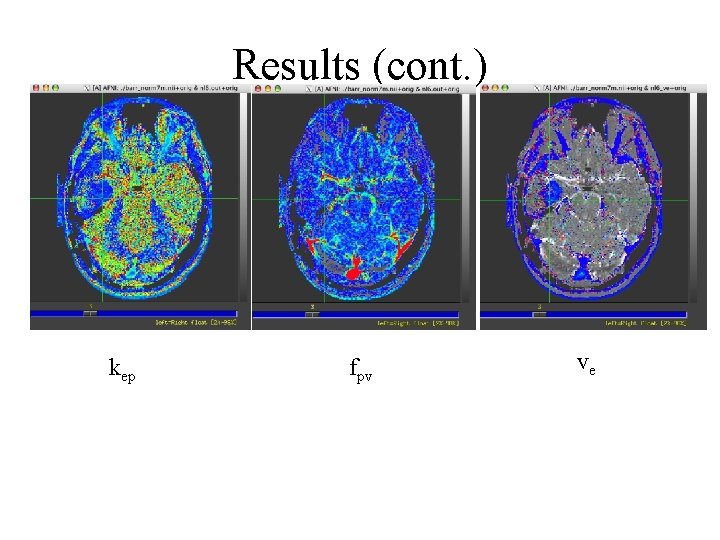
Results (cont. ) kep fpv ve

Discussion items: • R 1, i - function of fpv? • ve - limits, constraints on ktrans, kep • Powell optimization constraints • Mask voxel thresholds • Zero noise model speed • automatic sagittal sinus TAC versus manual, 3 d. ROIstats