Chromosome inversions in human populations Marta Ruiz Fernndez





- Slides: 23

Chromosome inversions in human populations Marta Ruiz Fernández Master in Advanced Genetics 17 December 2014

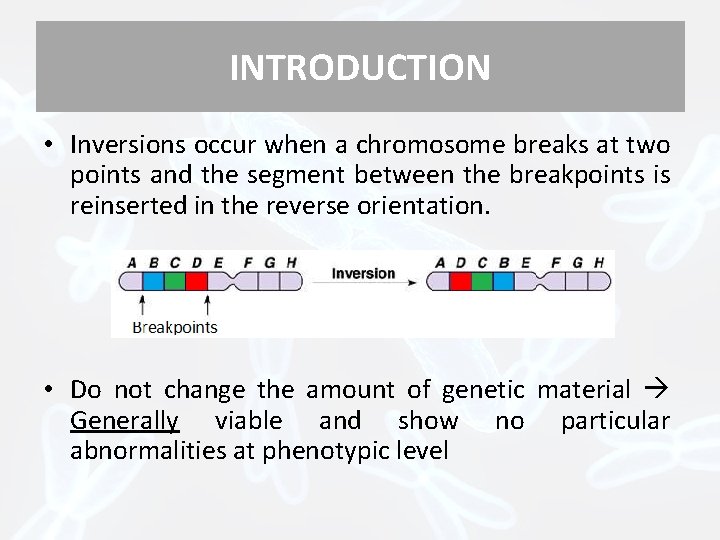
INTRODUCTION • Inversions occur when a chromosome breaks at two points and the segment between the breakpoints is reinserted in the reverse orientation. • Do not change the amount of genetic material Generally viable and show no particular abnormalities at phenotypic level

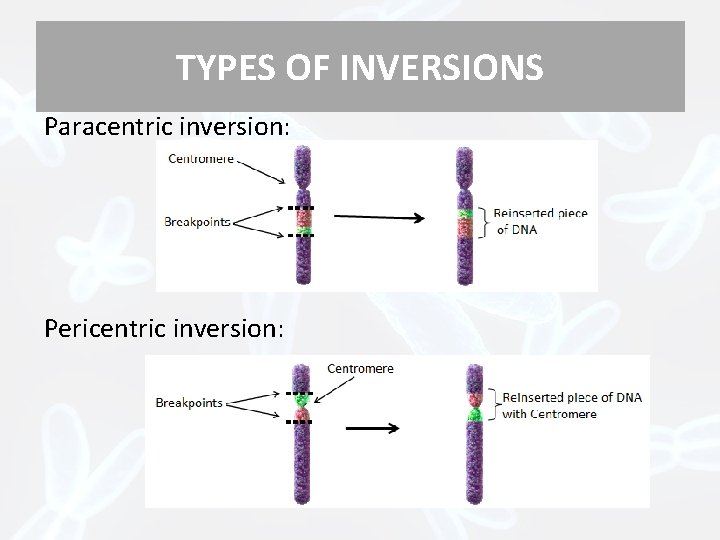
TYPES OF INVERSIONS Paracentric inversion: Pericentric inversion:

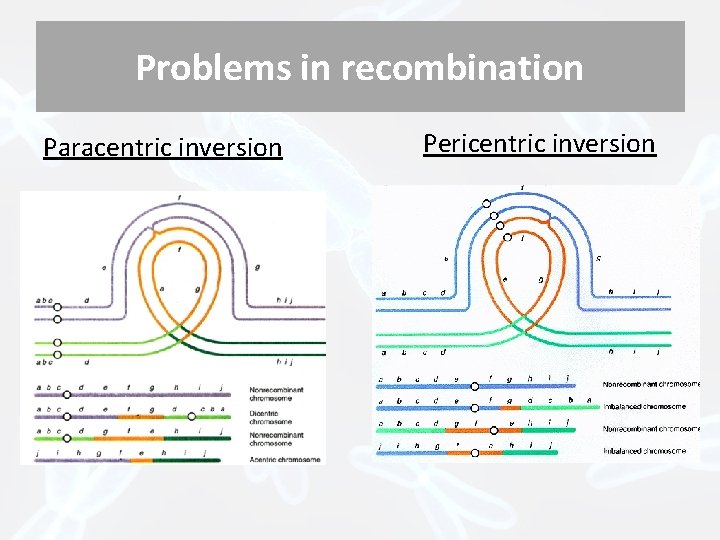
Problems in recombination Paracentric inversion Pericentric inversion

MECHANISMS OF INVERSION • Double strand break is needed for the inversion • “Hotspots” – Usually contains segmental duplications (SD) – Distance between SD = 10 -400 kb – 95 – 97% identity • Mechanisms – Non-allelic homologous recombination (NAHR) – Non-homologous end joining (NHEJ) – Fork stalling and template switching

CONSEQUENCES OF INVERSIONS • Recombination suppressed in heterozygotes – Reducing cross-over within inverted regions – If recombination occurs Not viable cells • Inversions could lead to insertions or deletions • Depending on the breakpoints position: – – Gene disruption Alter gene expression Generate new splicing sites Gene fusion NEUTRAL DELETERIOUS ADAPTATIVE

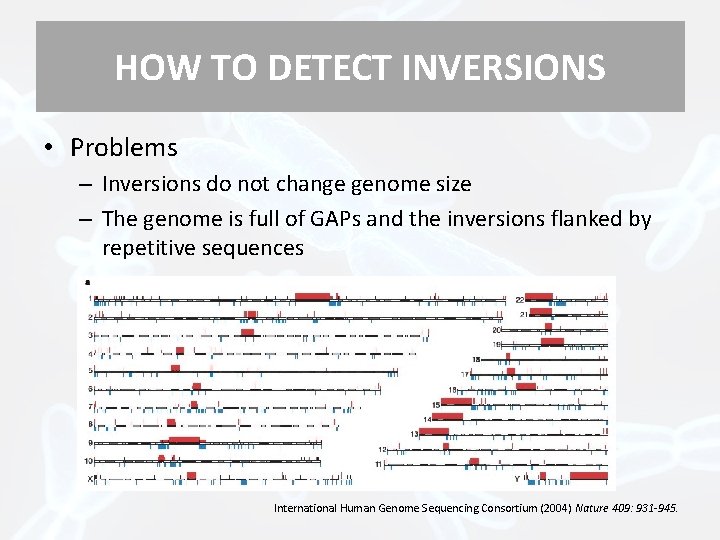
HOW TO DETECT INVERSIONS • Problems – Inversions do not change genome size – The genome is full of GAPs and the inversions flanked by repetitive sequences International Human Genome Sequencing Consortium (2004) Nature 409: 931 -945.

HOW TO DETECT INVERSIONS • Problems – Inversions do not change genome size – The genome is full of GAPs and the inversions flanked by repetitive sequences • Methods – Cytogenetic (conventional, FISH-based assay) – Pair-end sequencing – Genome assembly

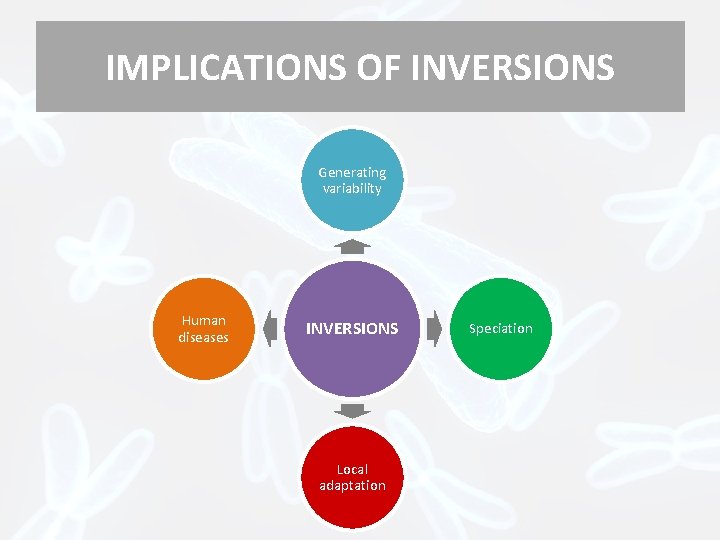
IMPLICATIONS OF INVERSIONS Generating variability Human diseases INVERSIONS Local adaptation Speciation

The role of inversions in human diversity • Hap. Map project Structural Variations

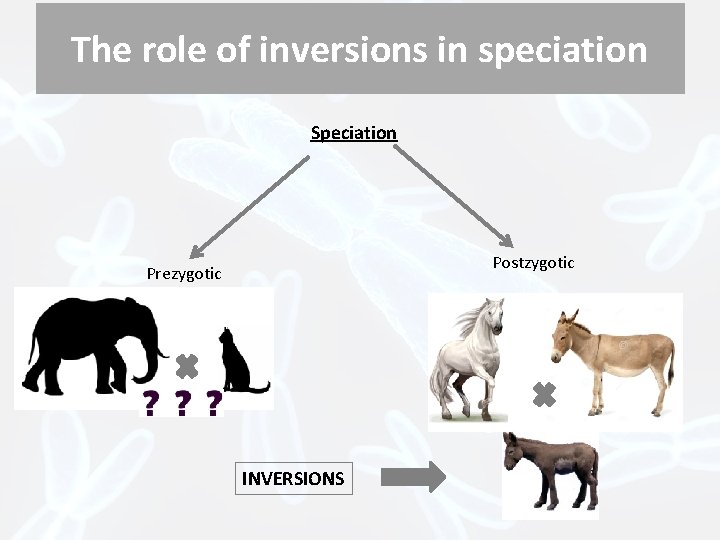
The role of inversions in speciation Speciation Postzygotic Prezygotic INVERSIONS

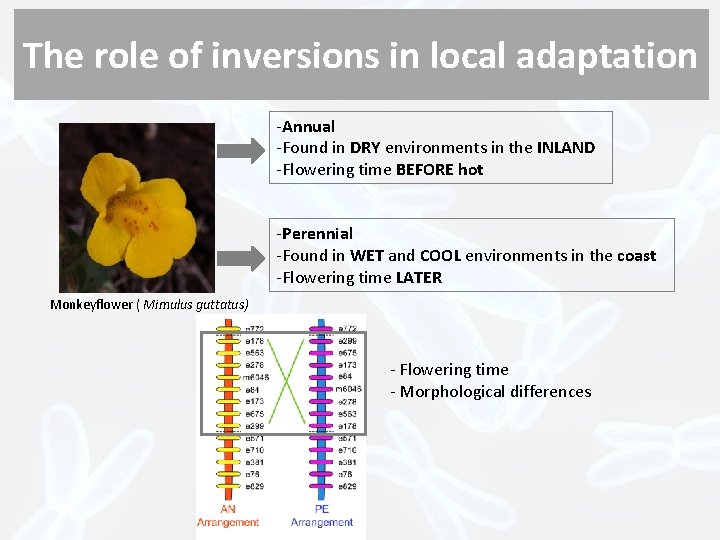
The role of inversions in local adaptation -Annual -Found in DRY environments in the INLAND -Flowering time BEFORE hot -Perennial -Found in WET and COOL environments in the coast -Flowering time LATER Monkeyflower ( Mimulus guttatus) - Flowering time - Morphological differences

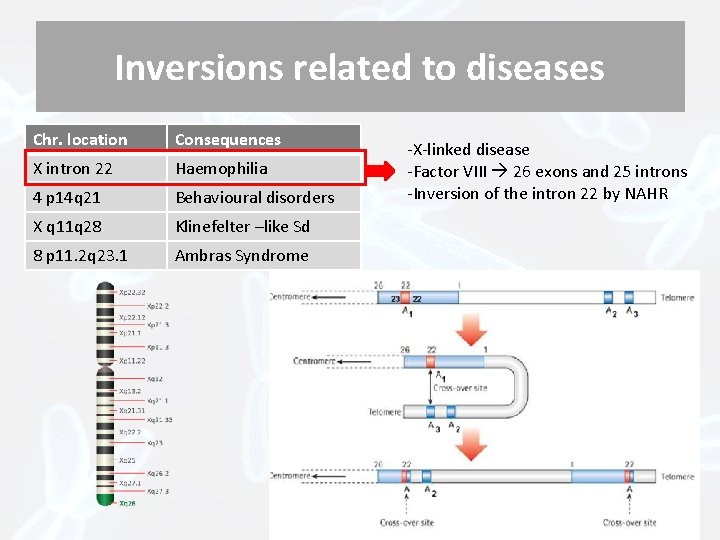
Inversions related to diseases Chr. location Consequences X intron 22 Haemophilia 4 p 14 q 21 Behavioural disorders X q 11 q 28 Klinefelter –like Sd 8 p 11. 2 q 23. 1 Ambras Syndrome -X-linked disease -Factor VIII 26 exons and 25 introns -Inversion of the intron 22 by NAHR

CONCLUSIONS • Several studies have reported many inversions • Way to generate variation between and within species • Diversity of effects depending on: – Where? (Coding/Non-coding, introns, exons) – Is any gene affected? – Does the inversion imply another chromosomal arrangement?

REFERENCES • Antonacci F, Kidd JM, Marques-Bonet T, Ventura M, Siswara P, Jiang Z, et al. Characterization of six human disease-associated inversion polymorphisms. Hum Mol Genet. 2009; 18: 2555– 66. • Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. Pathogenetics. 2008; 1: 4. • Bansal V, Bashir A, Bafna V. Evidence for large inversion polymorphisms in the human genome from Hap. Map data 2007: 219– 30. • Lee J, Han K, Meyer TJ, Kim H-S, Batzer M a. Chromosomal inversions between human and chimpanzee lineages caused by retrotransposons. PLo. S One. 2008; 3: e 4047. • Kirkpatrick M. How and why chromosome inversions evolve. PLo. S Biol. 2010; 8. • Lowry DB, Willis JH. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLo. S Biol. 2010; 8.

Chromosome inversions in human populations Marta Ruiz Fernández Master in Advanced Genetics 17 December 2014


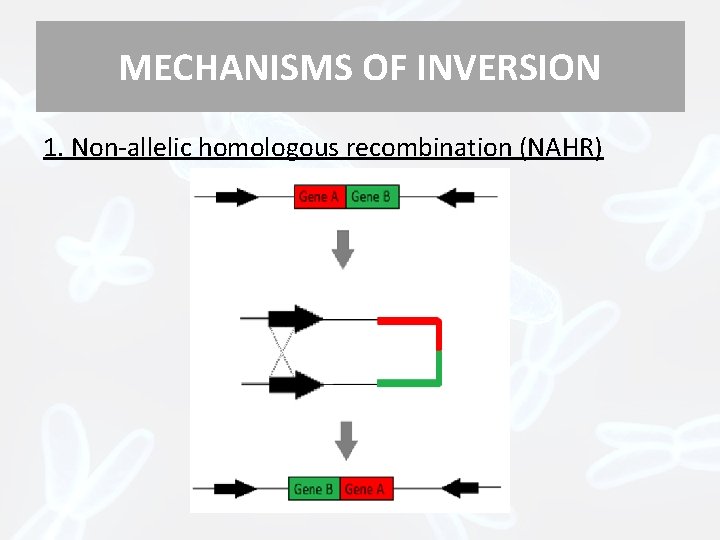
MECHANISMS OF INVERSION 1. Non-allelic homologous recombination (NAHR)

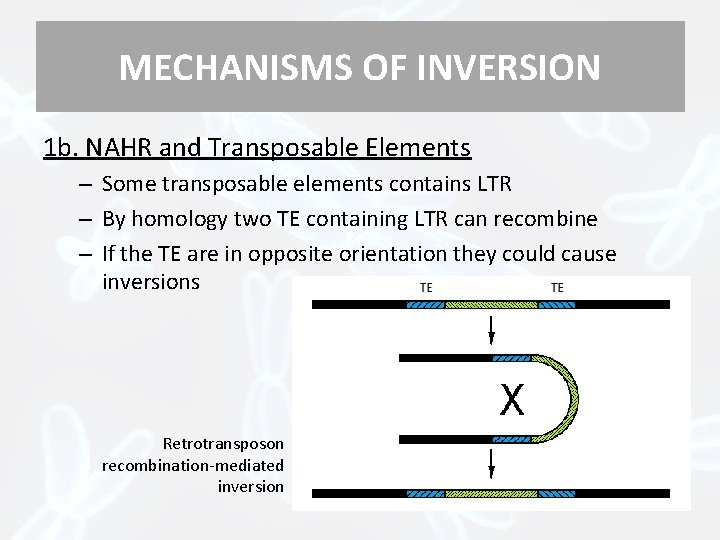
MECHANISMS OF INVERSION 1 b. NAHR and Transposable Elements – Some transposable elements contains LTR – By homology two TE containing LTR can recombine – If the TE are in opposite orientation they could cause inversions Retrotransposon recombination-mediated inversion

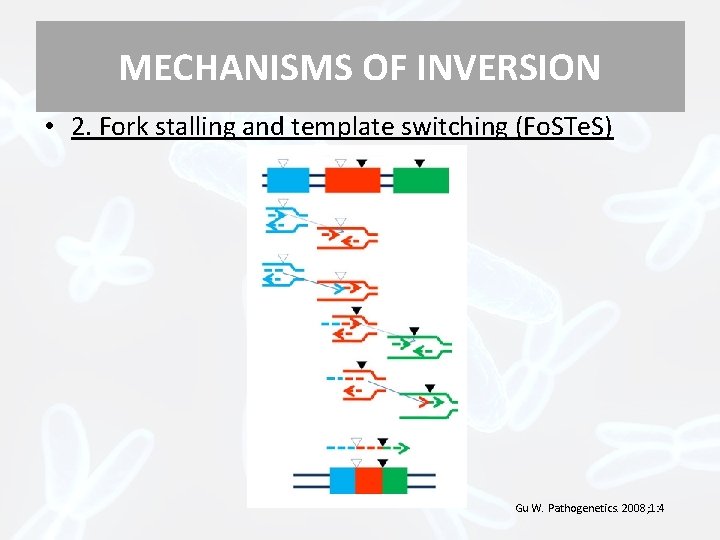
MECHANISMS OF INVERSION • 2. Fork stalling and template switching (Fo. STe. S) Gu W. Pathogenetics. 2008; 1: 4

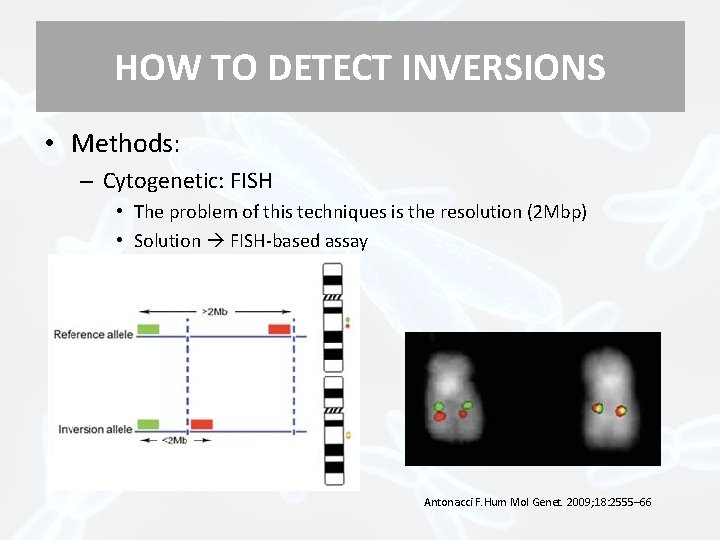
HOW TO DETECT INVERSIONS • Methods: – Cytogenetic: FISH • The problem of this techniques is the resolution (2 Mbp) • Solution FISH-based assay Antonacci F. Hum Mol Genet. 2009; 18: 2555– 66

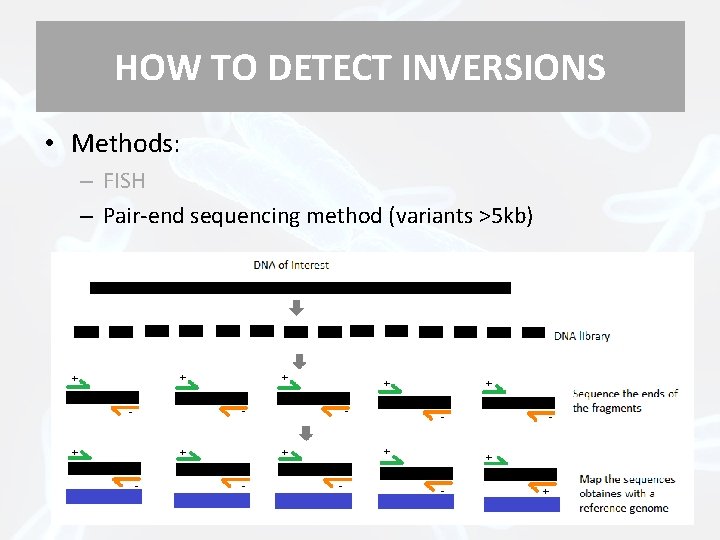
HOW TO DETECT INVERSIONS • Methods: – FISH – Pair-end sequencing method (variants >5 kb) + + - + - +

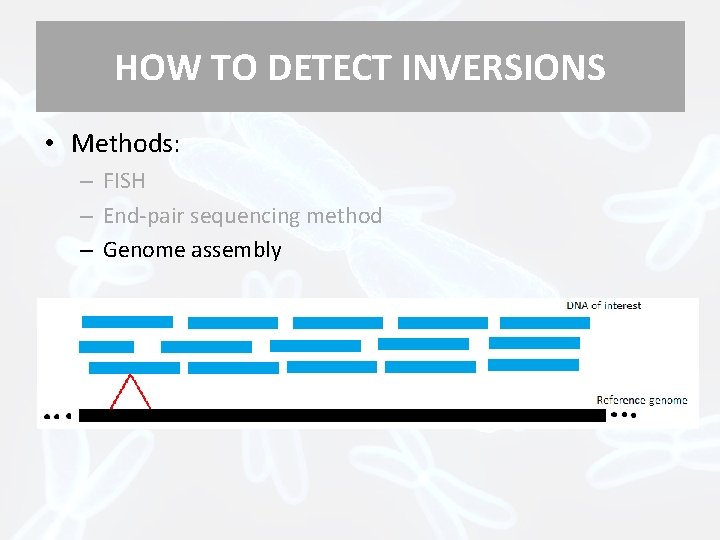
HOW TO DETECT INVERSIONS • Methods: – FISH – End-pair sequencing method – Genome assembly
 Alejandro fernndez
Alejandro fernndez Alejandro fernndez
Alejandro fernndez How to read chromosome
How to read chromosome Inversions 80s
Inversions 80s Inversions with negative adverbials
Inversions with negative adverbials Temperature inversions work to trap pollution when *
Temperature inversions work to trap pollution when * 7th chord inversion symbols
7th chord inversion symbols Divide and conquer counting inversions
Divide and conquer counting inversions Counting inversions divide and conquer
Counting inversions divide and conquer Inversion in english
Inversion in english Major 2
Major 2 Chromosome number
Chromosome number Carolina ruiz wpi
Carolina ruiz wpi Dr norman ruiz castaneda
Dr norman ruiz castaneda Daniel ruiz corona
Daniel ruiz corona Jess ruiz
Jess ruiz Ruiz ramos eliana
Ruiz ramos eliana Manuel ruiz vida
Manuel ruiz vida Nercida de ruiz
Nercida de ruiz Kubizm hermetyczny
Kubizm hermetyczny Bibliografia de carlos ruiz zafon
Bibliografia de carlos ruiz zafon Berta ruiz
Berta ruiz Rosario ortega ruiz
Rosario ortega ruiz Seppet
Seppet