Analyzing Promoter Sequences with Multilayer Perceptrons Glenn Walker





- Slides: 15

Analyzing Promoter Sequences with Multilayer Perceptrons Glenn Walker ECE 539

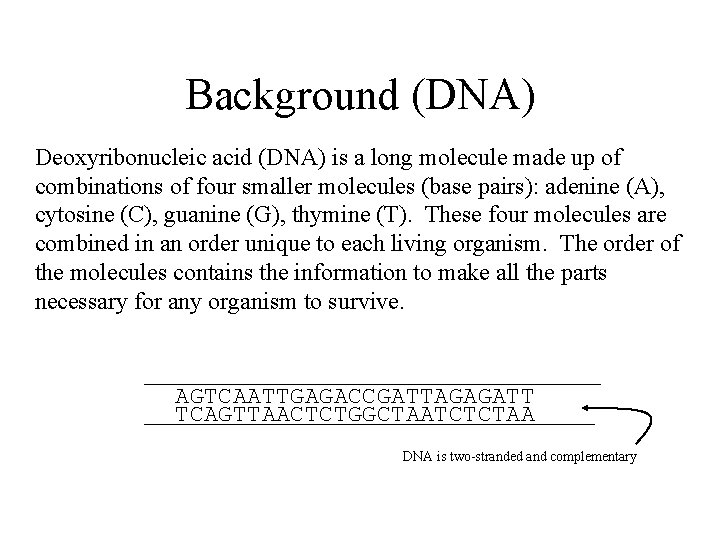
Background (DNA) Deoxyribonucleic acid (DNA) is a long molecule made up of combinations of four smaller molecules (base pairs): adenine (A), cytosine (C), guanine (G), thymine (T). These four molecules are combined in an order unique to each living organism. The order of the molecules contains the information to make all the parts necessary for any organism to survive. AGTCAATTGAGACCGATTAGAGATT TCAGTTAACTCTGGCTAATCTCTAA DNA is two-stranded and complementary

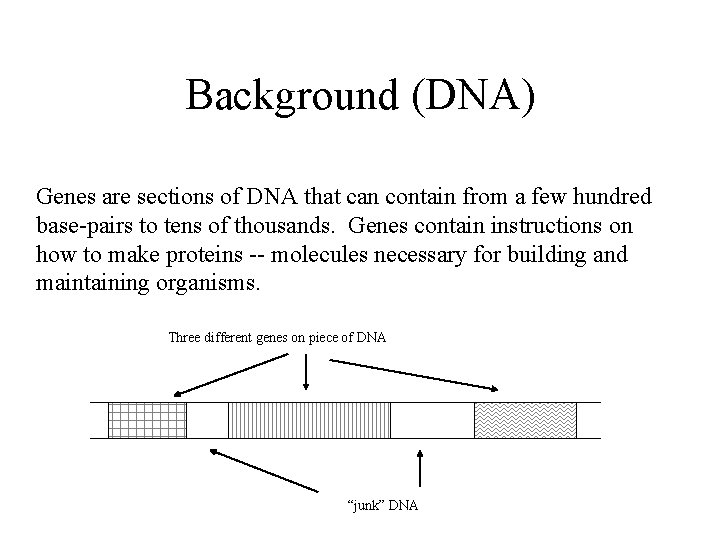
Background (DNA) Genes are sections of DNA that can contain from a few hundred base-pairs to tens of thousands. Genes contain instructions on how to make proteins -- molecules necessary for building and maintaining organisms. Three different genes on piece of DNA “junk” DNA

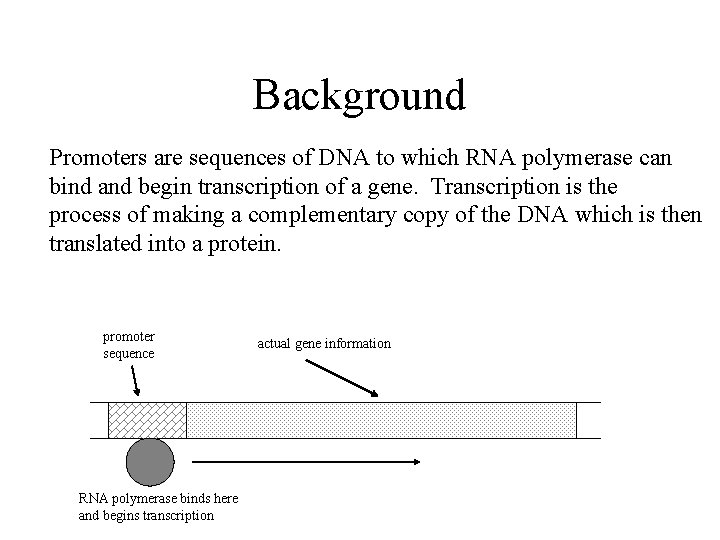
Background Promoters are sequences of DNA to which RNA polymerase can bind and begin transcription of a gene. Transcription is the process of making a complementary copy of the DNA which is then translated into a protein. promoter sequence RNA polymerase binds here and begins transcription actual gene information

Problem • Knowing gene locations is desirable for medical reasons • One way to find genes is to look for promoter regions • How do we find promoter regions?

One Solution • Promoter regions are highly conserved -different regions often contain similar patterns • We can train neural networks to recognize promoter regions • We choose a multilayer perceptron

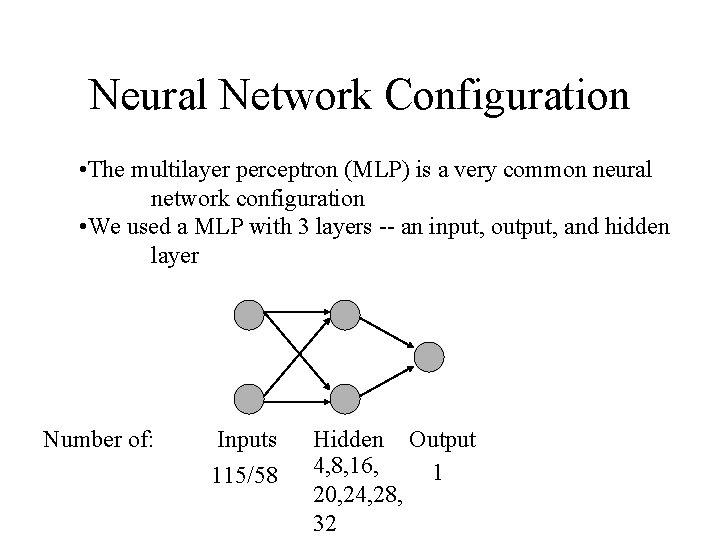
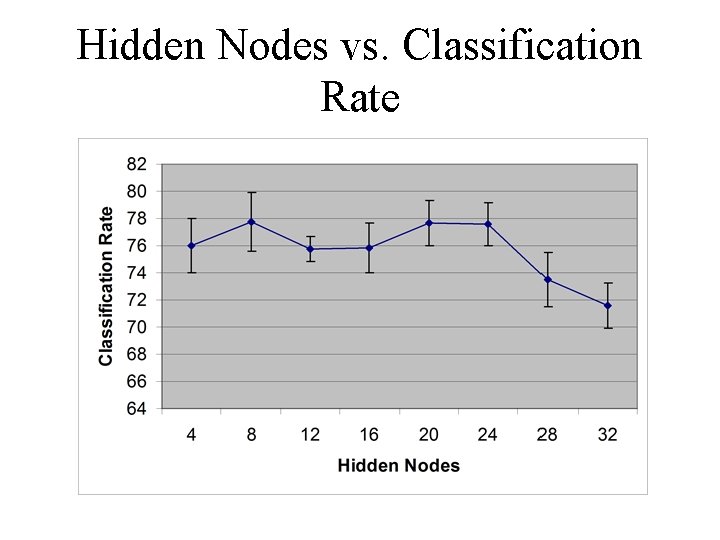
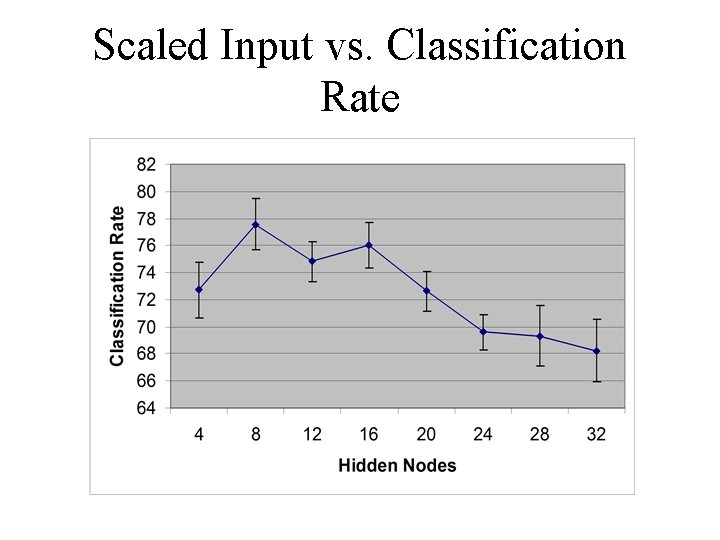
Neural Network Configuration • The multilayer perceptron (MLP) is a very common neural network configuration • We used a MLP with 3 layers -- an input, output, and hidden layer Number of: Inputs 115/58 Hidden Output 4, 8, 16, 1 20, 24, 28, 32

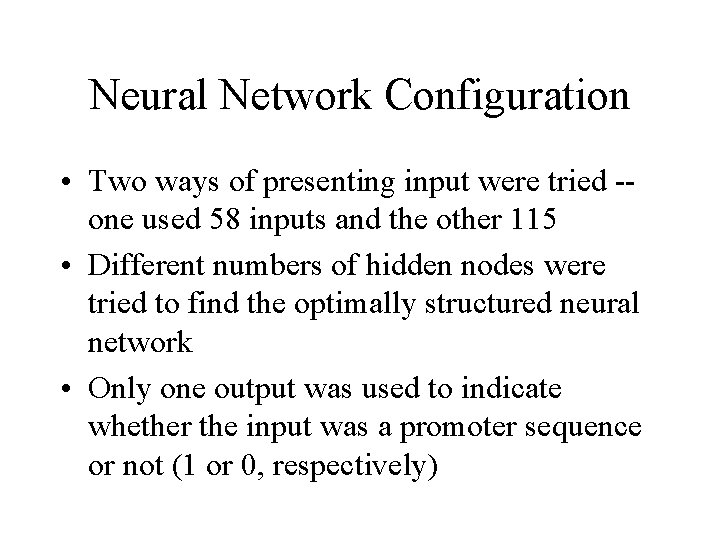
Neural Network Configuration • Two ways of presenting input were tried -one used 58 inputs and the other 115 • Different numbers of hidden nodes were tried to find the optimally structured neural network • Only one output was used to indicate whether the input was a promoter sequence or not (1 or 0, respectively)

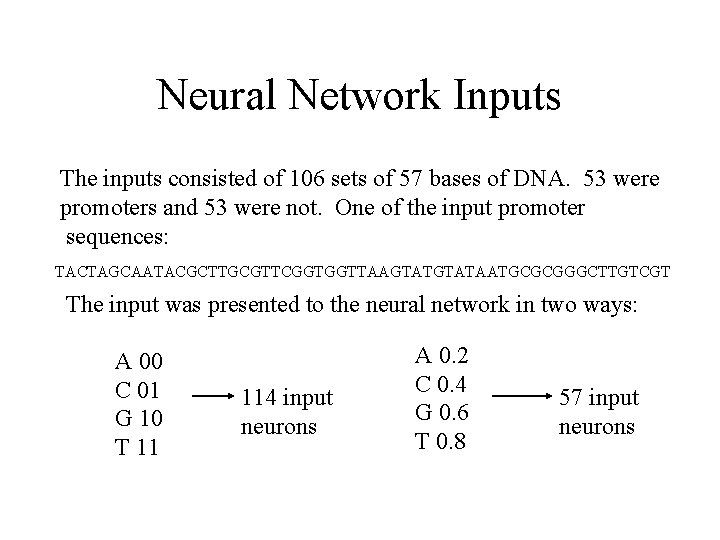
Neural Network Inputs The inputs consisted of 106 sets of 57 bases of DNA. 53 were promoters and 53 were not. One of the input promoter sequences: TACTAGCAATACGCTTGCGTTCGGTGGTTAAGTATAATGCGCGGGCTTGTCGT The input was presented to the neural network in two ways: A 00 C 01 G 10 T 11 114 input neurons A 0. 2 C 0. 4 G 0. 6 T 0. 8 57 input neurons

Neural Network Training Each configuration was run 10 times. Within each of the 10 runs, 106 runs were performed. For each of these, 105 of the promoter sequences were used for training with the 106 th used for testing. The testing sequences were changed for each of the 106 runs so that each sequence was the test sequence only once. Ten runs were necessary since weights for the MLP were initialized to random values which might have led to different classifications for the same input sequence.

Hidden Nodes vs. Classification Rate

Scaled Input vs. Classification Rate

Compared to Others Walker (NN) O’Neil (NN) Towell (KBANN) O’Neil (Rule-based) ID 3 (Decision tree) 78% 83% > 90% 76%

Conclusion • Not the best but not the worst • Using a hybrid technique would improve results • The MLP is a very useful tool for the field of bioinformatics

References Harley, C. B. and Reynolds, R. P. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Research, 15(5): 2343 -2361. O’Neill, M. C. 1991. Training back-propagation neural networks to define and detect DNA-binding sites. Nucleic Acids Research, 19(2): 313 -318. Quinlan, J. 1986. Induction of decision trees. Machine Learning, 1: 81 -106. Towell, G. G. , Shavlik, J. W. , and Noordewier, M. O. 1990. Refinement of Approximate Domain Theories by Knowledge-Based Neural Networks. AAAI-90, 861 -866.