Use of FISH in sequencing tomato chromosome 6

- Slides: 14

Use of FISH in sequencing tomato chromosome 6 René Klein Lankhorst Hans de Jong Korea meeting 2007

Topics • FISH to define the borders between euchromatin and heterochromatin • FISH to target novel seed BACs towards BAC-oceans

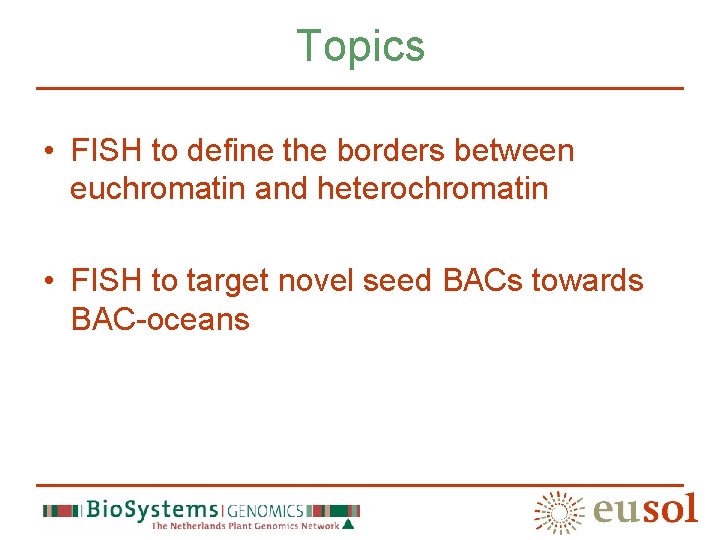
Major repeats of the tomato genome

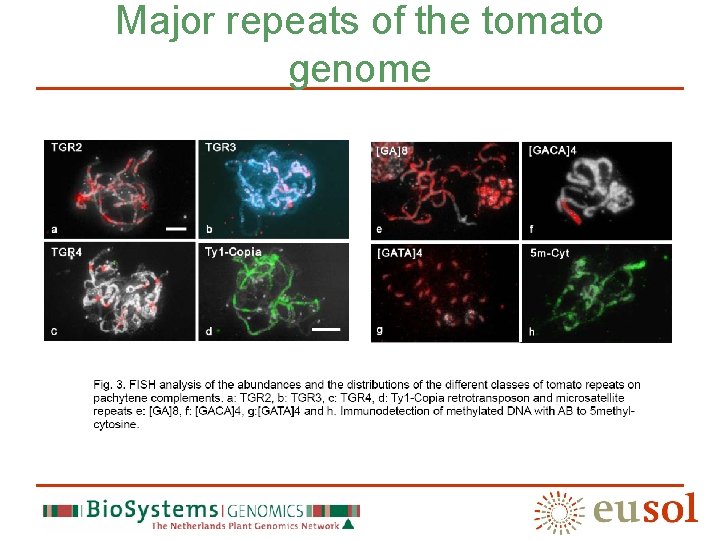
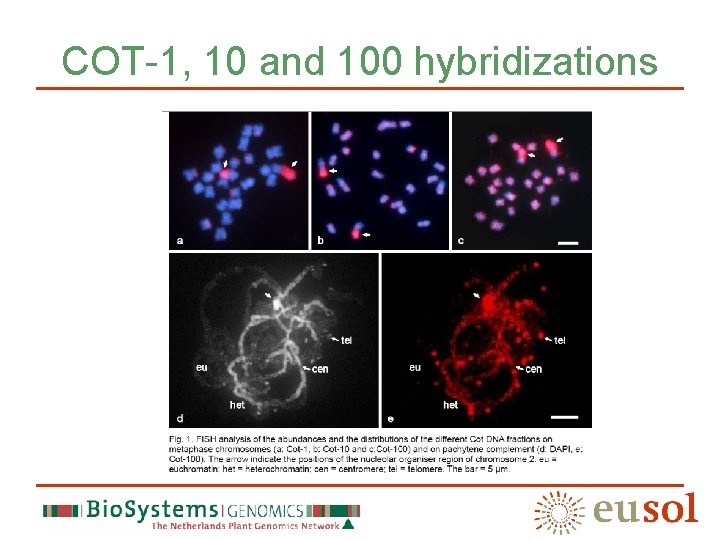
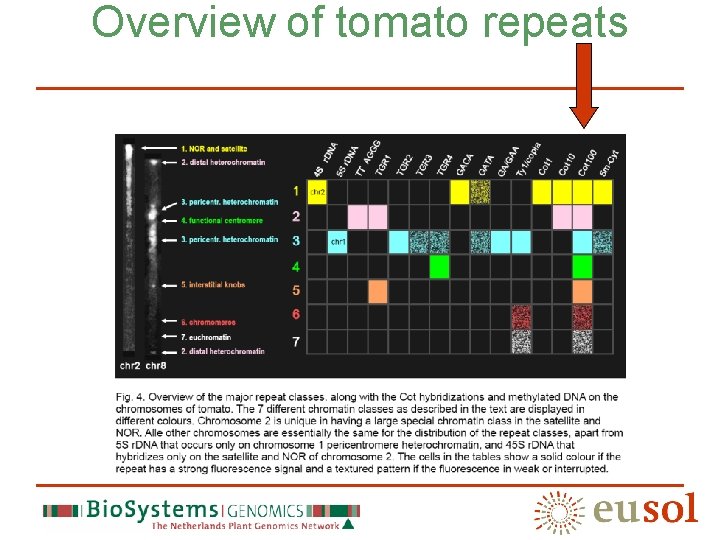
Seven chromatin classes class Repeats (abundant) NOR/satellite 45 S r. DNA, GACA, GATA, Cot 1 -10, Methylated DNA Distal heterochromatin TTTAGGG, TGR 1, Cot 100 Pericentromere heterochromatin TGR 2, TGR 3, TGR 4, GATA, GA/GAA, Ty 1/Copia, Cot 100, Methylated DNA Centromere TGR 4, Cot 100 Interstitial knob TGR 1, Cot 100 Chromomere Ty 1/copia, Cot 100 (weak) Euchromatin Cot 100 (weak)

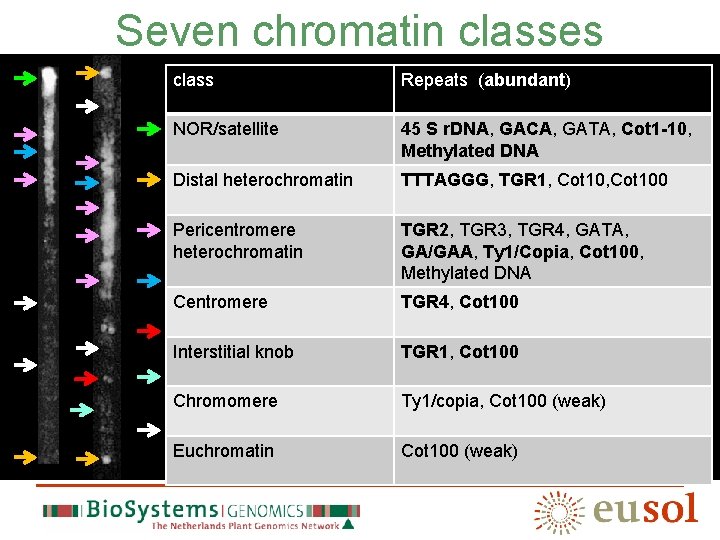
COT-1, 10 and 100 hybridizations

Overview of tomato repeats

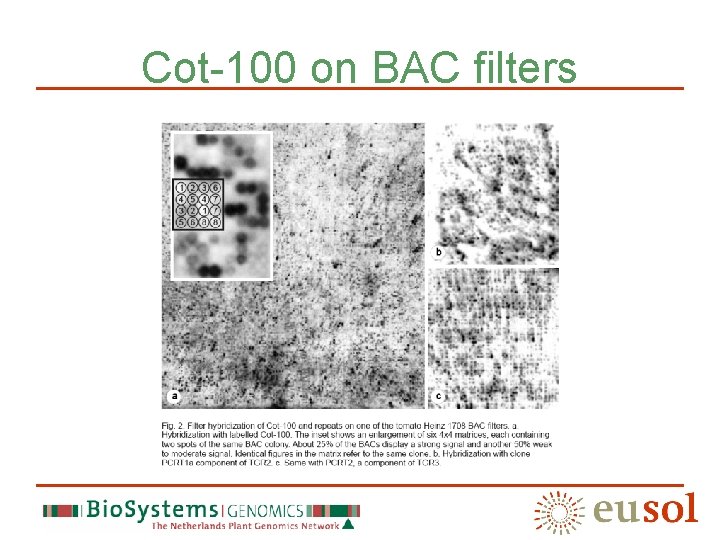
Cot-100 on BAC filters

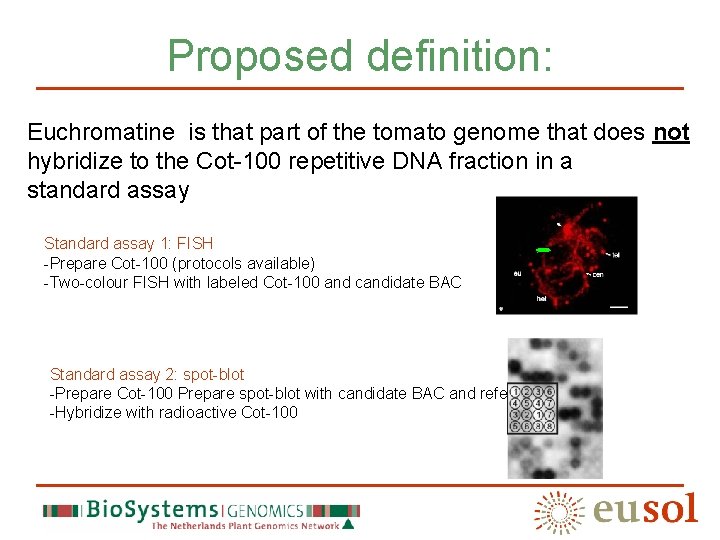
Proposed definition: Euchromatine is that part of the tomato genome that does not hybridize to the Cot-100 repetitive DNA fraction in a standard assay Standard assay 1: FISH -Prepare Cot-100 (protocols available) -Two-colour FISH with labeled Cot-100 and candidate BAC Standard assay 2: spot-blot -Prepare Cot-100 Prepare spot-blot with candidate BAC and reference BACs -Hybridize with radioactive Cot-100

Part 2 • FISH to target novel seed BACs towards BAC-oceans

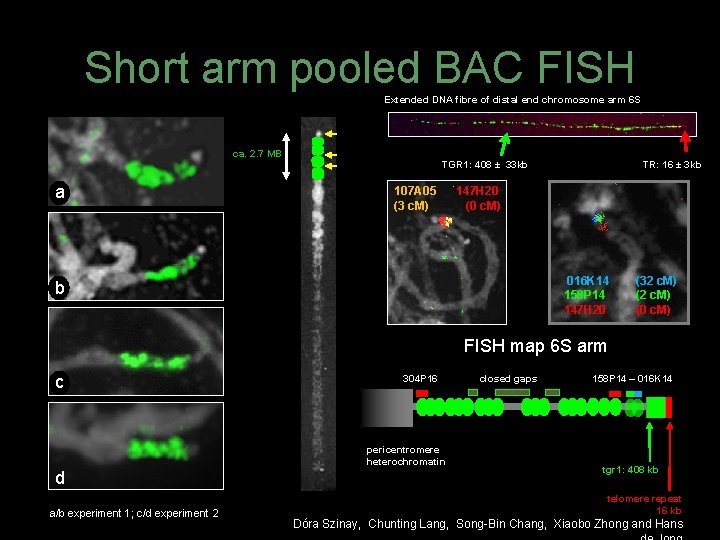
Short arm pooled BAC FISH Extended DNA fibre of distal end chromosome arm 6 S ca. 2. 7 MB a TGR 1: 408 ± 33 kb 107 A 05 (3 c. M) TR: 16 ± 3 kb 147 H 20 (0 c. M) 016 K 14 158 P 14 147 H 20 b (32 c. M) (0 c. M) FISH map 6 S arm c 304 P 16 pericentromere heterochromatin d a/b experiment 1; c/d experiment 2 closed gaps 158 P 14 – 016 K 14 tgr 1: 408 kb telomere repeat 16 kb Dóra Szinay, Chunting Lang, Song-Bin Chang, Xiaobo Zhong and Hans

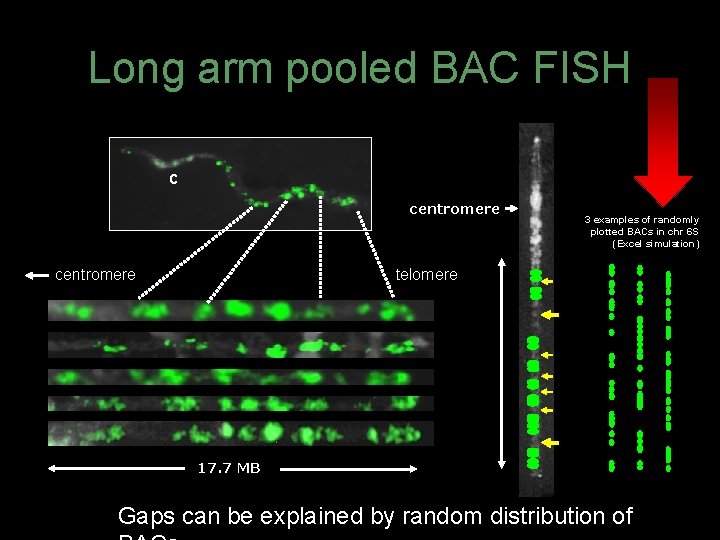
Long arm pooled BAC FISH c centromere 3 examples of randomly plotted BACs in chr 6 S (Excel simulation) telomere 17. 7 MB Gaps can be explained by random distribution of

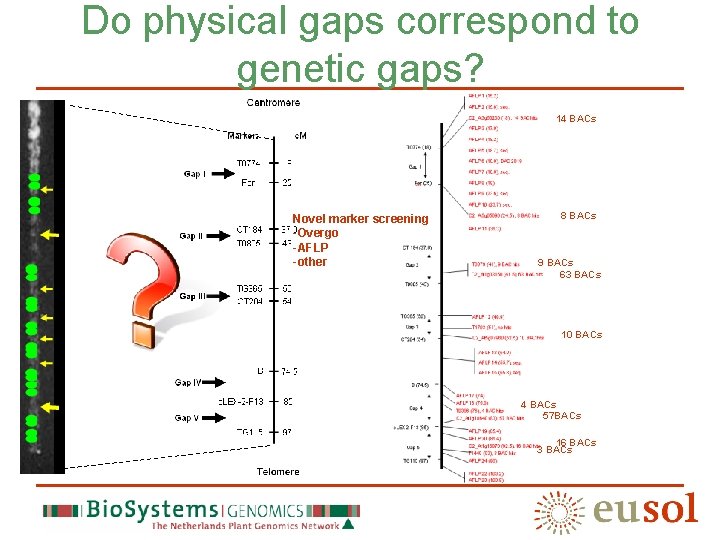
Do physical gaps correspond to genetic gaps? 14 BACs Novel marker screening -Overgo -AFLP -other 8 BACs 9 BACs 63 BACs 10 BACs 4 BACs 57 BACs 16 BACs 3 BACs

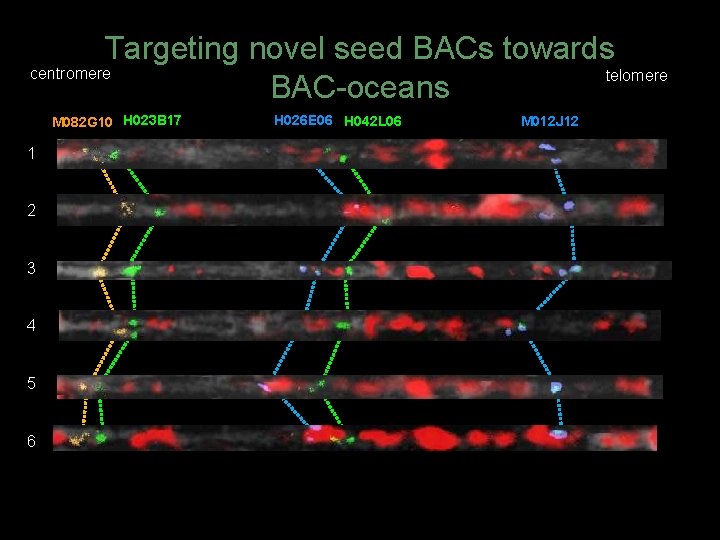
Targeting novel seed BACs towards centromere telomere BAC-oceans M 082 G 10 H 023 B 17 1 2 3 4 5 6 H 026 E 06 H 042 L 06 M 012 J 12

Take-home messages • Cot-100 can be used to discriminate between euchromatin and heterochromatin and thus can be used to define borders. • (For the long arm of chr. 6) genetic gaps and physical gaps correspond and thus genetic information can be used to target novel seed-BACs towards BAC-oceans • (For the long arm of chr. 6) no indication for a bias in the Hind. III library nor in the F 2. 2000 genetic map was found.
 One fish two fish red fish blue fish ride
One fish two fish red fish blue fish ride One fish, two fish, blowfish, blue fish
One fish, two fish, blowfish, blue fish Characteristics of actinopterygii
Characteristics of actinopterygii How to read chromosome
How to read chromosome Cartilaginous fish reproduction
Cartilaginous fish reproduction Cartilaginous fish vs bony fish
Cartilaginous fish vs bony fish A big fish swims up and swallows a small fish at rest
A big fish swims up and swallows a small fish at rest Cut evenly from large slabs of frozen fillets
Cut evenly from large slabs of frozen fillets Tomato breeding objectives
Tomato breeding objectives Parasitism
Parasitism Every pale tomato slice
Every pale tomato slice Sabyonne
Sabyonne Is a moss living or nonliving
Is a moss living or nonliving Soul tomato skyrim
Soul tomato skyrim Sifat kimia bahan neutral
Sifat kimia bahan neutral