Tomato Chromosome 4 A Mapping Sequencing Update Christine




- Slides: 17

Tomato Chromosome 4: A Mapping & Sequencing Update Christine Nicholson Mapping Core Group Welcome Trust Sanger Institute, UK 28 th September 2005

FPC Database • Mapping strategy = to develop the physical map in order to select minimal tiling paths across the chromosome to sequence, in conjunction with BES data and markers. • FPC database worked on in-house at Sanger • Obtained from Arizona Genomics Institute • The FPC Database: – – – LE_HBa library fingerprints 88, 584 fingerprinted BACs (68% of total library) Possibly due to contamination of library Therefore, Have ~ 10 X coverage in the fingerprint map.

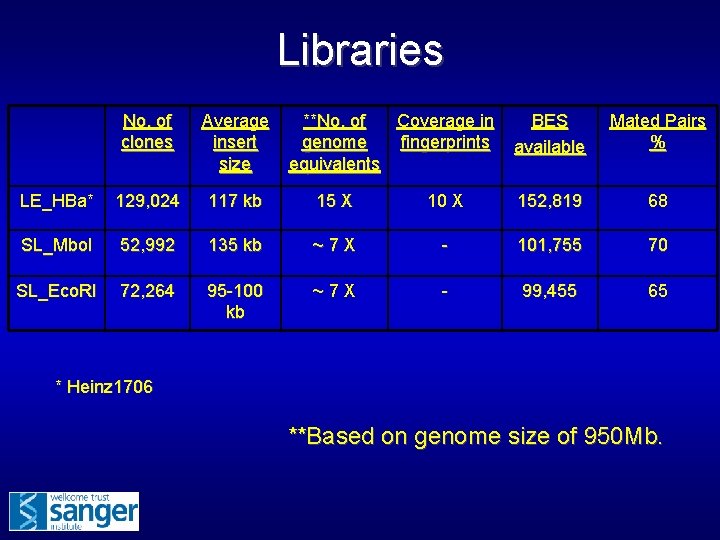
Libraries No. of clones Average insert size **No. of Coverage in genome fingerprints equivalents BES available Mated Pairs % LE_HBa* 129, 024 117 kb 15 X 10 X 152, 819 68 SL_Mbo. I 52, 992 135 kb ~7 X - 101, 755 70 SL_Eco. RI 72, 264 95 -100 kb ~7 X - 99, 455 65 * Heinz 1706 **Based on genome size of 950 Mb.

Chromosome 4 Contigs • Examined seed BACs/contigs for each chromosome using overgo probes. (Conducted by Cornell) ftp: //ftp. sgn. cornell. edu/tomato_genome/seedbacs/20050112_chr 4_long_short. xls • 54 markers on chromosome 4 in the current FPC build • Each potential chromosome 4 contig assessed in silico in FPC: - structure based on fingerprints - marker content – assigned to how many contigs? - possible merges to other contigs? September 2005: 58 contigs currently on chr 4

Pilot Sequencing • • • Select 5 BACs to form “pilot sequencing”. In two different regions of the chromosome Tomato BACs are being processed through our sequencing pipeline. • Apply our Finishing programmes to tomato clones & examine sequence features e. g. repeats. Sequencing Pipeline at WTSI……

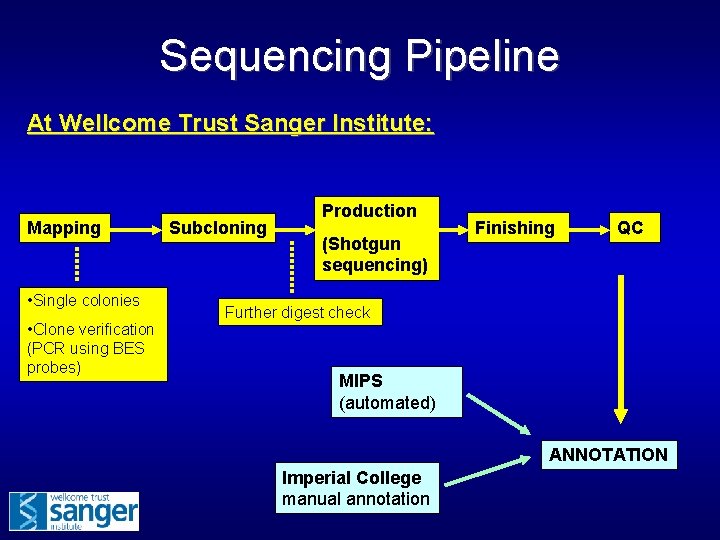
Sequencing Pipeline At Wellcome Trust Sanger Institute: Mapping • Single colonies • Clone verification (PCR using BES probes) Subcloning Production (Shotgun sequencing) Finishing QC Further digest check MIPS (automated) ANNOTATION Imperial College manual annotation

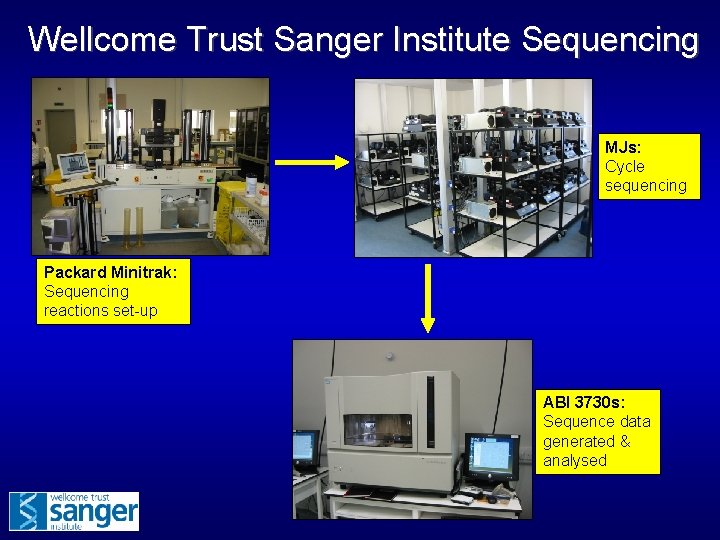
Wellcome Trust Sanger Institute Sequencing MJs: Cycle sequencing Packard Minitrak: Sequencing reactions set-up ABI 3730 s: Sequence data generated & analysed

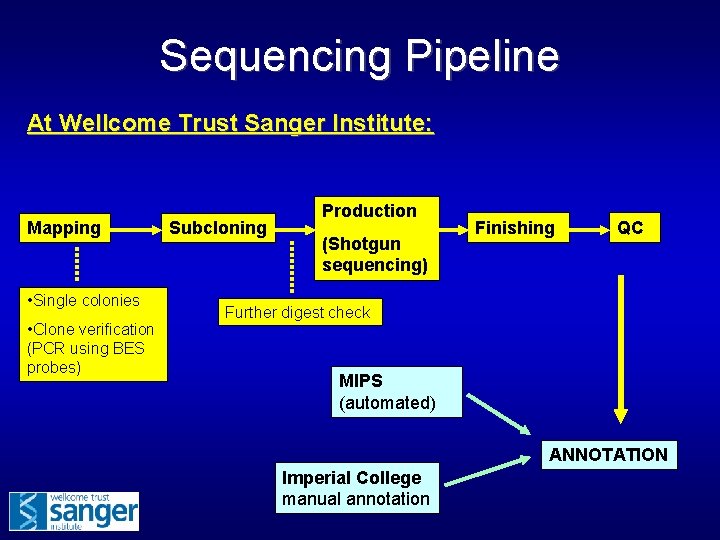
Sequencing Pipeline At Wellcome Trust Sanger Institute: Mapping • Single colonies • Clone verification (PCR using BES probes) Subcloning Production (Shotgun sequencing) Finishing QC Further digest check MIPS (automated) ANNOTATION Imperial College manual annotation

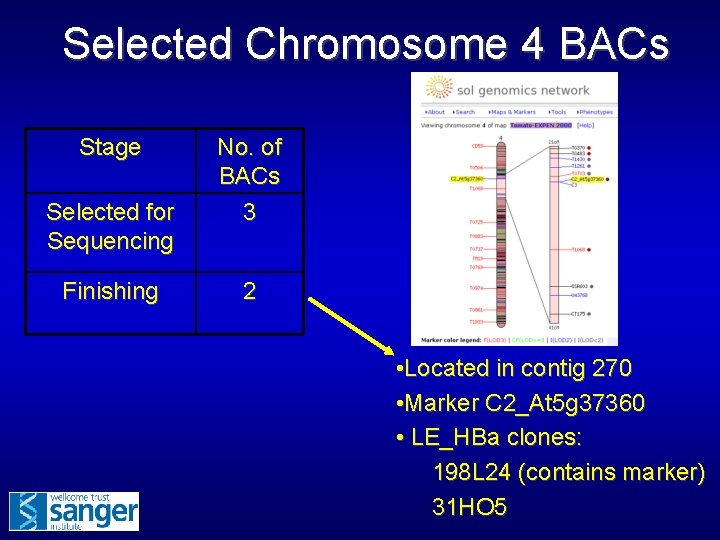
Selected Chromosome 4 BACs Stage Selected for Sequencing Finishing No. of BACs 3 2 • Located in contig 270 • Marker C 2_At 5 g 37360 • LE_HBa clones: 198 L 24 (contains marker) 31 HO 5

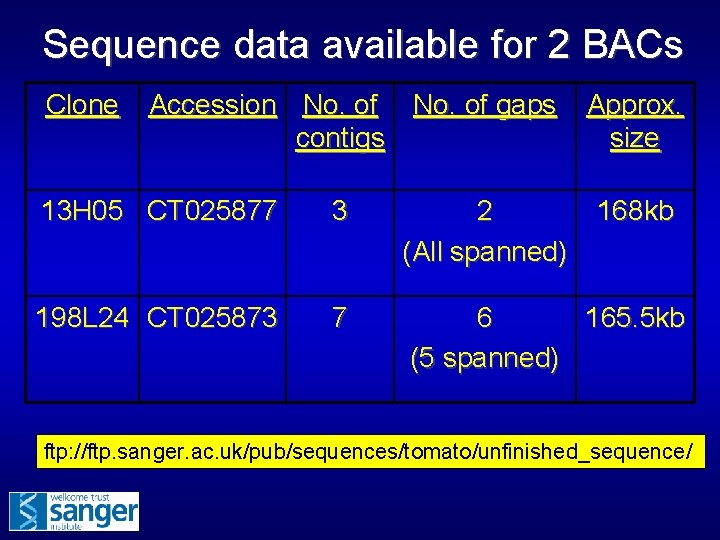
Sequence data available for 2 BACs Clone Accession No. of gaps contigs Approx. size 13 H 05 CT 025877 3 2 (All spanned) 168 kb 198 L 24 CT 025873 7 6 165. 5 kb (5 spanned) ftp: //ftp. sanger. ac. uk/pub/sequences/tomato/unfinished_sequence/

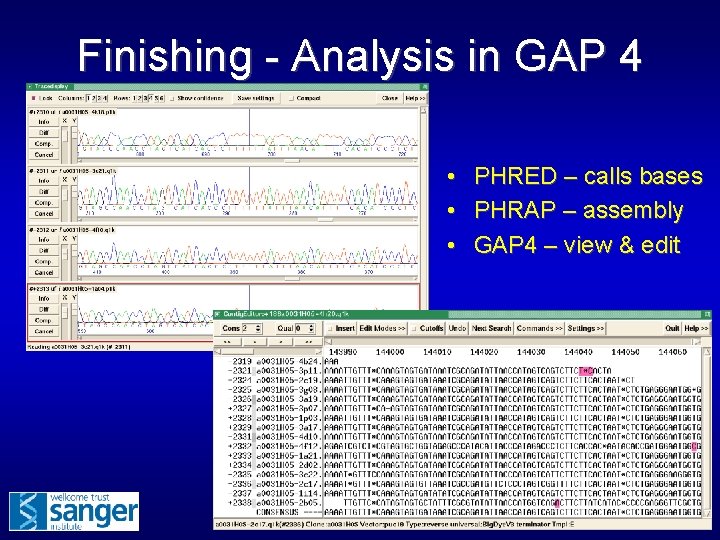
Finishing - Analysis in GAP 4 • • • PHRED – calls bases PHRAP – assembly GAP 4 – view & edit

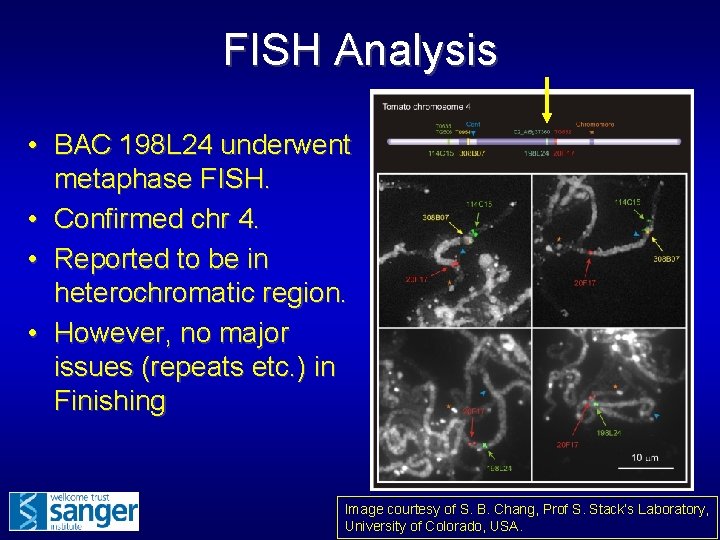
FISH Analysis • BAC 198 L 24 underwent metaphase FISH. • Confirmed chr 4. • Reported to be in heterochromatic region. • However, no major issues (repeats etc. ) in Finishing Image courtesy of S. B. Chang, Prof S. Stack’s Laboratory, University of Colorado, USA.

Genes in Sequenced BACs? • Used WUBLASTX align proteins against the tomato sequence. • Partial gene highlighted in each BAC 31 H 05 putative carboxyl-terminal peptidase 198 L 24 AMT 1. 2 Ammonium transporter 1 member 2

Mapping Strategy * AIM = Reduce the current contig number * WHY? • Select longer minimal tilepaths across the chromosome • Smaller overlaps of selected sequence BACs (aim for 1520 kb) • More efficient sequencing HOW? • Work on FPC database to improve continuity • Walk off sequenced clones (once available) using BES hits • Incorporate further BES/fingerprint data as generated • Possible walk from contig ends by hybridization.

Further Map Development & Fingerprinting? • Have been able to replicate the fingerprinting technique from AGI. • 31 H 05 and 198 L 24 fingerprints confirmed. • Will use this for future clone verification. • Currently have 10 X coverage in fingerprints. • Investigating the practicalities and possibilities of augmenting the fingerprint database → SL_Mbo. I library?

Further BAC Selection Strategy • Intend to select minimal tilepaths to sequence ideally over reduced number of contigs on chromosome 4. • Will FISH more BACs across the chromosome obtain confirmation of chromosome location of BACs & contigs they are contained within.

Acknowledgements Wellcome Trust Sanger Institute: Jane Rogers Sean Humphray Carol Scott Karen Barlow Helen Beasley Sarah Sims Jennifer Harrow Carol Carder Paul Hunt Mark Maddison Imperial College London: Gerard Bishop University of Warwick: Graham Seymour FUNDING Cornell University: Lukas Mueller Arizona Genomics Institute: Rod Wing Seunghee Lee Colorado State University: Stephen Stack Song-Bin Chang Scottish Crop Research Institute: Glenn Bryan