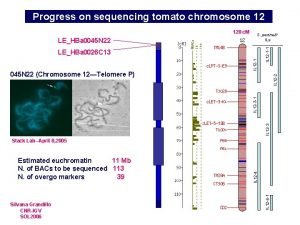
Progress in the sequencing of tomato chromosome 12

- Slides: 7

Progress in the sequencing of tomato chromosome 12 (See also poster number 39) The project is funded by the Italian Government (FIRB and Agronanotech projects) and by the European Union (EU-SOL project). Three main units are involved: ENEA, Rome (Prof. Giuliano) University of Naples (Prof. Frusciante) University of Padua (Prof. Valle) PAG 2008, San Diego

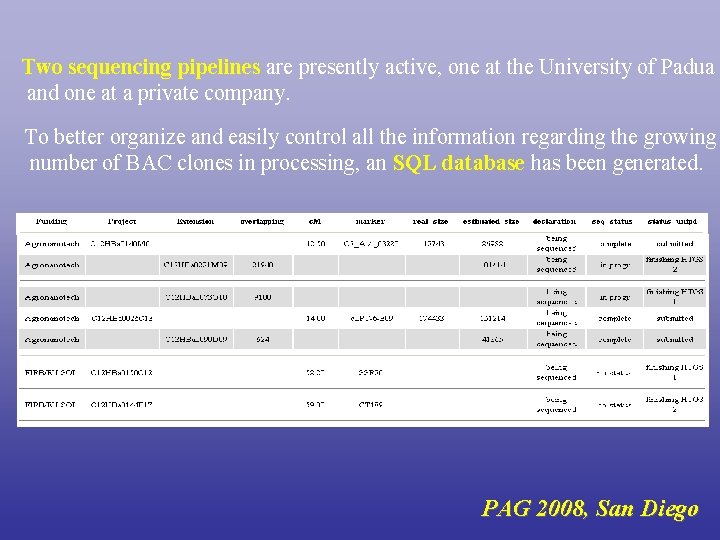
Two sequencing pipelines are presently active, one at the University of Padua and one at a private company. To better organize and easily control all the information regarding the growing number of BAC clones in processing, an SQL database has been generated. PAG 2008, San Diego

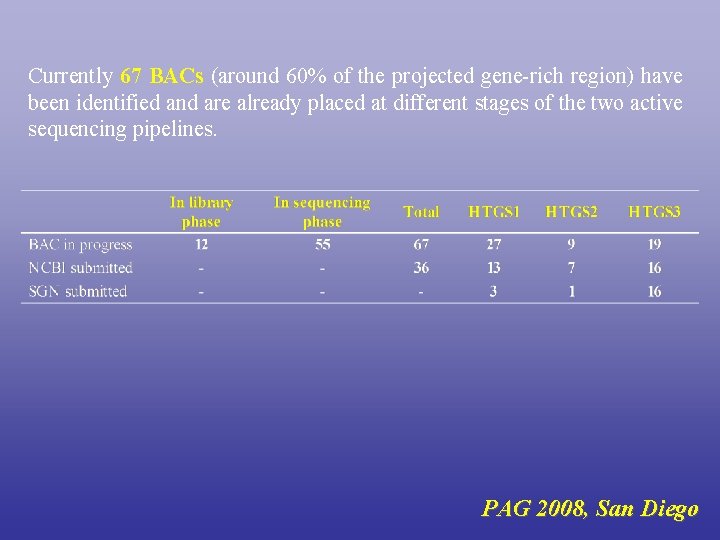
Currently 67 BACs (around 60% of the projected gene-rich region) have been identified and are already placed at different stages of the two active sequencing pipelines. PAG 2008, San Diego

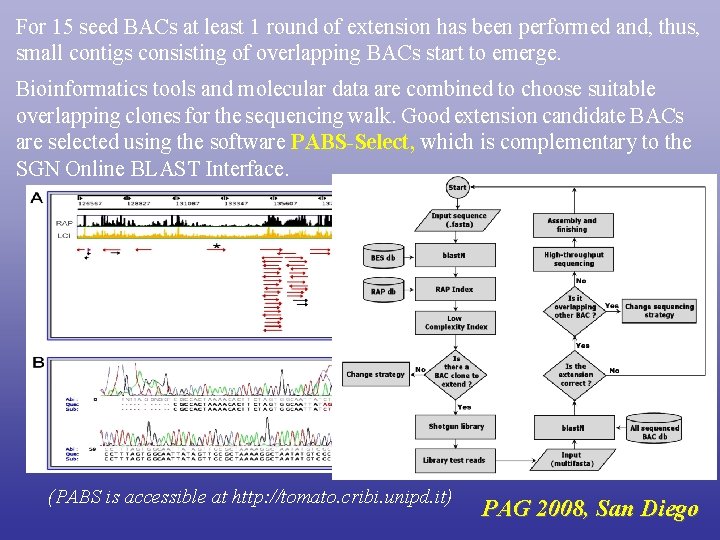
For 15 seed BACs at least 1 round of extension has been performed and, thus, small contigs consisting of overlapping BACs start to emerge. Bioinformatics tools and molecular data are combined to choose suitable overlapping clones for the sequencing walk. Good extension candidate BACs are selected using the software PABS-Select, which is complementary to the SGN Online BLAST Interface. (PABS is accessible at http: //tomato. cribi. unipd. it) PAG 2008, San Diego

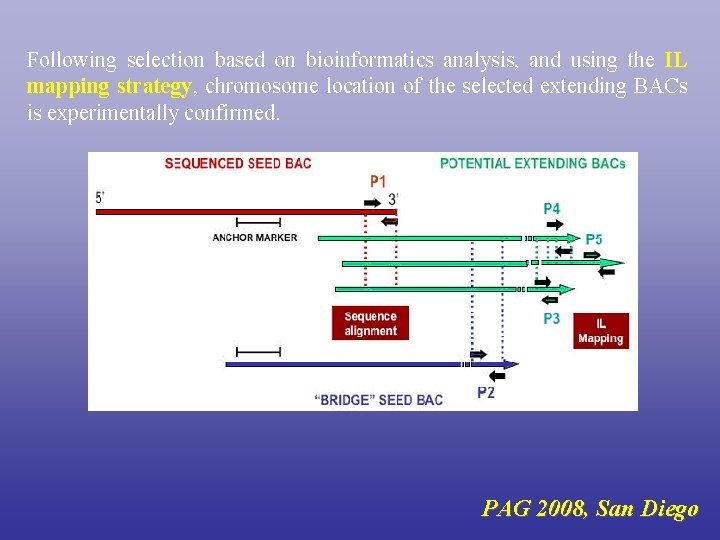
Following selection based on bioinformatics analysis, and using the IL mapping strategy, chromosome location of the selected extending BACs is experimentally confirmed. PAG 2008, San Diego

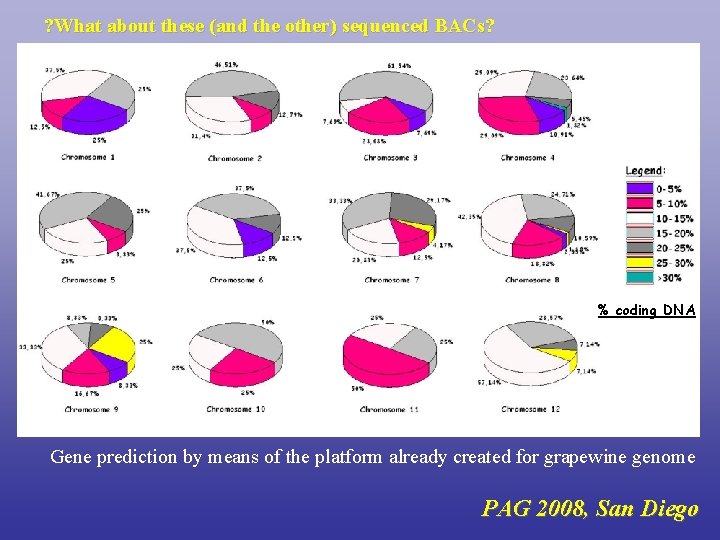
? What about these (and the other) sequenced BACs? % coding DNA Gene prediction by means of the platform already created for grapewine genome PAG 2008, San Diego

Acknowledgments ENEA - Rome Giovanni Giuliano Elio Fantini Giulia Falcone Marco Pietrella CNR-IGV, Portici (Naples) Silvana Grandillo CRIBI & Univ. of Padua Giorgio Valle University of Naples ‘Federico II’ (Dept. DISSPA & DSFB) Luigi Frusciante Amalia Barone Mara Ercolano Maria Luisa Chiusano Alessandro Vezzi Sara Todesco Riccardo Schiavon Silvia Pescarolo Alessandro Zambon Michela D’Angelo Rosa Paparo Walter Sanseverino Sara Torre PAG 2008, San Diego