Topic 16 Lecture 22 Species and Speciation What















- Slides: 39

Topic 16. Lecture 22. Species and Speciation What is species? Species is a key concept used to describe biodiversity. Indeed, Darwin's book was called "The Origin of Species". So, what is species? So, far we treated this concept simplistically, saying that a species is a set of similar individuals, and if all individuals within a set are similar, tightly related, connected, and compatible to each other, and, at the same time, dissimilar, only distantly related, disconnected, and incompatible to all other individuals, we are dealing with a "good species". Obviously, common ancestry of different modern "species" implies that there could be no good species if we take into account all organisms, modern and ancient. If we are willing to consider only modern organisms, sometimes we encounter reasonably good species.

Still, it is obvious that gradual evolution is inconsistent with all species being good. It does not make sense to ask when exactly independent evolution of two lineages makes them different species. How many grains constitute a heap? (one of Zeno's paradoxes). Exactly how many years, after the Isthmus of Panama has been formed, did it take for isolated populations to become different species? - A stupid question.

Indeed, very often modern species are not "good", and do not have definite boundaries. On the one hand , connected forms can be incompatible. Two reproductively isolated forms are connected by a continuous "bridge" consisting of perfectly fit intermediate forms. On the other hand, disconnected forms can be compatible. Two rhododendrons, Rhododendron catawbiense (North America, left) and R. fortunei (China, right) produce fertile hybrids (center).

Even within an apparently uniform "species", careful analysis often reveals some degree of incompatibility between individuals from different populations (outbreeding depression). Indeed, compatibility is not an all-or-nothing trait. For example, crosses between individuals from different populations of a copepod Tigriopus californicus result in backcross and F 2 hybrid breakdown for a variety of fitness related measures. The magnitude of this breakdown increases with evolutionary divergence between populations (J. of Evolutionary Biology 19, 2040, 2006). So are different populations of T. californicus different species? We must avoid asking such questions too seriously.

OK, so what is species? We can characterize diversity of life from many perspectives, and ask whether organisms from some set: 1) compete and interbreed with each other (if yes, they belong to the same population)? 2) are all similar to each other and different from other organisms (if yes, they belong to the same "form of life")? 3) are more closely related to each other than to any other organism (if yes, they belong to the same clade)? 4) are connected to each other (if yes, they belong to the same "cluster") 5) are compatible to each other (if yes, they belong to the same species). Aquilegia formosa A. pubescens I prefer to reserve the word species for consideration of compatibility and incompatibility between organisms. Different "species concepts" simply try to capture different aspects of biodiversity, and it is important to avoid terminological confusions.

Using word "species" for consideration of compatibility and incompatibility is known as "biological species concept", which defines species as "a set of actually and potentially interbreeding populations, reproductively isolated from other such sets" - a common definition of BSC. This definition makes sense, if we understand interbreeding as a test for compatibility (testing compatibility of asexuals is difficult, although the concept can be applied to them, too). Still, is not flexible enough to describe reality (also, I prefer to consider individuals, and not populations): 1) What if we find only a mild degree of reproductive isolation (outbreeding depression)? 2) What to do if transitivity is violated? Indeed, in some cases A can interbreed with B, B can interbreed with C, but A and C are incompatible. Transitivity (A = B and B = C imply A = C), along with reflexivity (A = A) and symmetry (A = B implies B = A) is necessary to have good, non-overlapping classes. Trouble is, Nature does not always cooperate.

It is better to think in terms of conspecificity of pairs of organisms ("binary", instead of "unary" definition of species): two individuals belong to the same species to the extent to which they are compatible to each other, in the sense that any organism with a "mixed" genome will be fit. Speciation is evolutionary origin of individual that are incompatible to their ancestors. Every two humans are 100% (or nearly so) the same species, and a human and an elephant are 100% different species. 100, 000 years of independent evolution - still the same species 100, 000 years of independent evolution - a different species 500, 000 years of independent evolution. We might eventually know if a H. sapiens can have healthy kids with a H. neanderthalensis. If we have a set of individuals such that every two ones from the set are fully compatible to each other, and each one is incompatible to anybody else, we have a "good species".

However, good species appear to be more of an exception than the norm, and there is no usually need to argue whether two individuals "really" belong to the same species, if they are in a grey area. Occasionally, this issue becomes important legally, but not scientifically, due to Endangered Species Act. A proposal to list polar bears as protected avoids the issue of whether they are a species. Phylogenetically, polar bears are nested within brown bears, and they are probably compatible to each other, at least intrinsically.

So the really interesting issue is the origin of incompatibility between lineages that evolved from the common ancestor. Indeed, this may look like a paradox: how can natural selection (survival of the fittest) lead to low fitness of hybrids? It is possible for two incompatible species to evolve from the common ancestor without violating dictate of natural selection? The answer to the second question is certainly "yes": horse-donkey divergence never involved a mule phase! Still, the origin of incompatibility, or speciation, is a fascinating process, and it is now relatively wellunderstood, at least at the population level. Darwin already realized that incompatibility between different species “is not a specially endowed quality, but is incidental on other acquired differences, ” (Origin of Species, p. 245) and is caused by a hybrid's “organization having been disturbed by two organizations having been compounded into one” (p. 266).

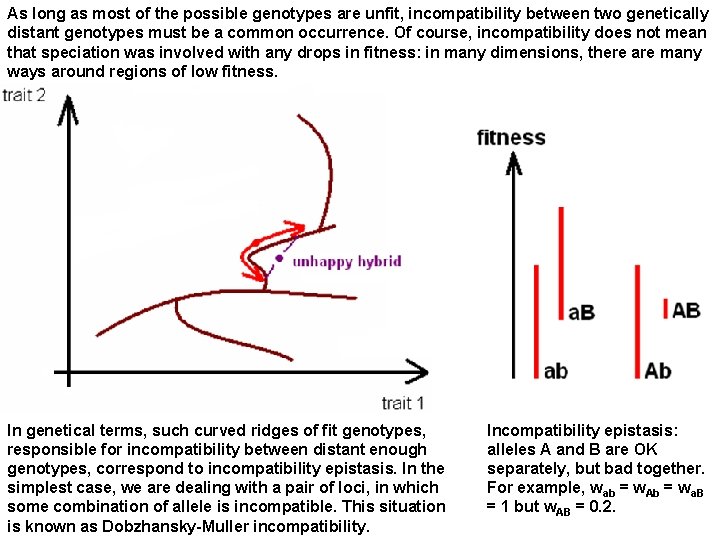
As long as most of the possible genotypes are unfit, incompatibility between two genetically distant genotypes must be a common occurrence. Of course, incompatibility does not mean that speciation was involved with any drops in fitness: in many dimensions, there are many ways around regions of low fitness. In genetical terms, such curved ridges of fit genotypes, responsible for incompatibility between distant enough genotypes, correspond to incompatibility epistasis. In the simplest case, we are dealing with a pair of loci, in which some combination of allele is incompatible. This situation is known as Dobzhansky-Muller incompatibility. Incompatibility epistasis: alleles A and B are OK separately, but bad together. For example, wab = w. Ab = wa. B = 1 but w. AB = 0. 2.

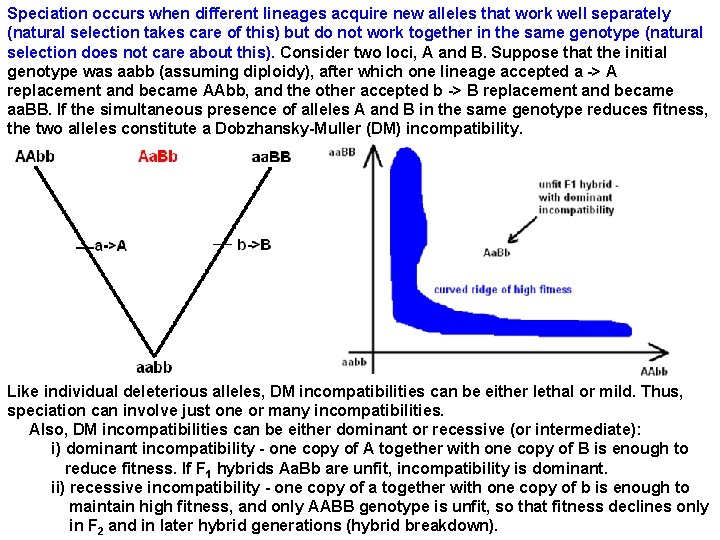
Speciation occurs when different lineages acquire new alleles that work well separately (natural selection takes care of this) but do not work together in the same genotype (natural selection does not care about this). Consider two loci, A and B. Suppose that the initial genotype was aabb (assuming diploidy), after which one lineage accepted a -> A replacement and became AAbb, and the other accepted b -> B replacement and became aa. BB. If the simultaneous presence of alleles A and B in the same genotype reduces fitness, the two alleles constitute a Dobzhansky-Muller (DM) incompatibility. Like individual deleterious alleles, DM incompatibilities can be either lethal or mild. Thus, speciation can involve just one or many incompatibilities. Also, DM incompatibilities can be either dominant or recessive (or intermediate): i) dominant incompatibility - one copy of A together with one copy of B is enough to reduce fitness. If F 1 hybrids Aa. Bb are unfit, incompatibility is dominant. ii) recessive incompatibility - one copy of a together with one copy of b is enough to maintain high fitness, and only AABB genotype is unfit, so that fitness declines only in F 2 and in later hybrid generations (hybrid breakdown).

Many DM incompatibilities are at least partially recessive, leading to "Haldane's Rule: "hybrid sterility or inviability tends to afflict the heterogametic sex more than the homogametic sex". Let as compare two cases: 1) Both A and B are autosomal loci 2) A is sex-linked and B is autosomal AAbb x aa. BB AAbb x a-BB | Aa. Bb A-Bb fit, if incompatibility is recessive daughters are fit, but sons are unfit In mammals and flies, males are the XY (heterogametic) sex, but in birds and butterflies females are heterogametic (ZW). In the first case, hybrid sons often have lower fitness than hybrid daughters, and the pattern is opposite in the other cases (there are only very few exceptions). A sterile female butterfly hybrid

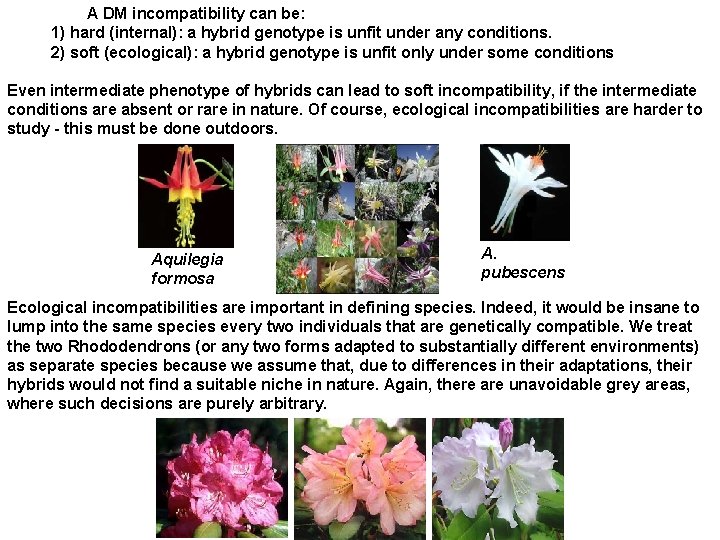
A DM incompatibility can be: 1) hard (internal): a hybrid genotype is unfit under any conditions. 2) soft (ecological): a hybrid genotype is unfit only under some conditions Even intermediate phenotype of hybrids can lead to soft incompatibility, if the intermediate conditions are absent or rare in nature. Of course, ecological incompatibilities are harder to study - this must be done outdoors. Aquilegia formosa A. pubescens Ecological incompatibilities are important in defining species. Indeed, it would be insane to lump into the same species every two individuals that are genetically compatible. We treat the two Rhododendrons (or any two forms adapted to substantially different environments) as separate species because we assume that, due to differences in their adaptations, their hybrids would not find a suitable niche in nature. Again, there are unavoidable grey areas, where such decisions are purely arbitrary.

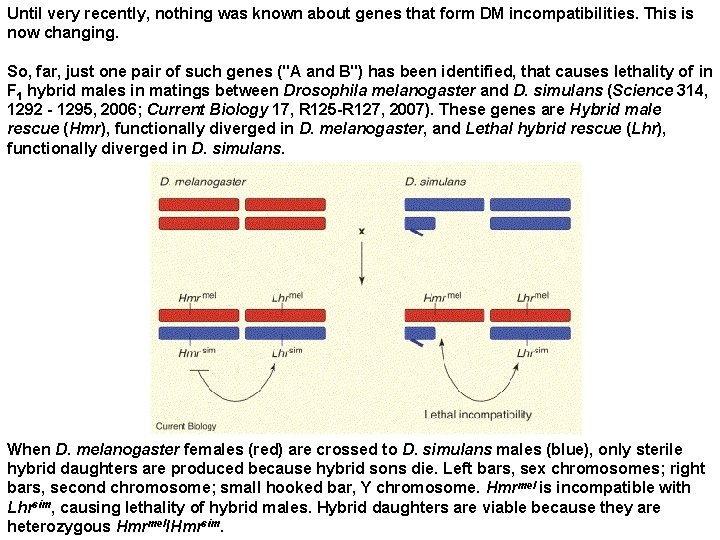
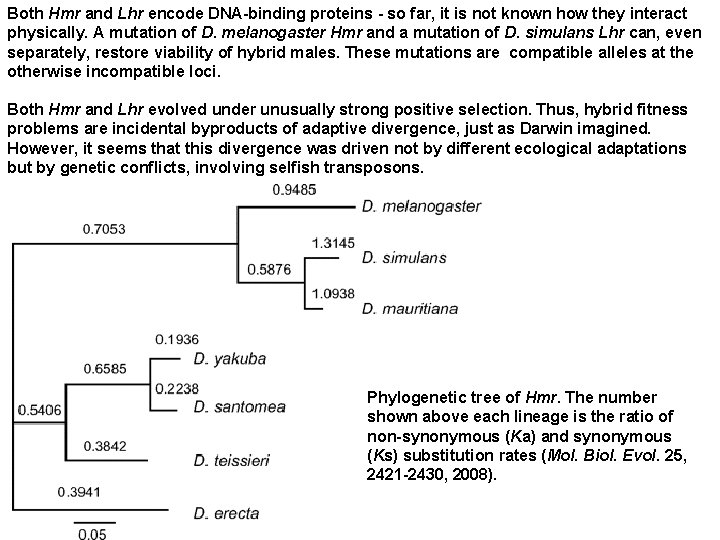
Until very recently, nothing was known about genes that form DM incompatibilities. This is now changing. So, far, just one pair of such genes ("A and B") has been identified, that causes lethality of in F 1 hybrid males in matings between Drosophila melanogaster and D. simulans (Science 314, 1292 - 1295, 2006; Current Biology 17, R 125 -R 127, 2007). These genes are Hybrid male rescue (Hmr), functionally diverged in D. melanogaster, and Lethal hybrid rescue (Lhr), functionally diverged in D. simulans. When D. melanogaster females (red) are crossed to D. simulans males (blue), only sterile hybrid daughters are produced because hybrid sons die. Left bars, sex chromosomes; right bars, second chromosome; small hooked bar, Y chromosome. Hmrmel is incompatible with Lhrsim, causing lethality of hybrid males. Hybrid daughters are viable because they are heterozygous Hmrmel/Hmrsim.

Both Hmr and Lhr encode DNA-binding proteins - so far, it is not known how they interact physically. A mutation of D. melanogaster Hmr and a mutation of D. simulans Lhr can, even separately, restore viability of hybrid males. These mutations are compatible alleles at the otherwise incompatible loci. Both Hmr and Lhr evolved under unusually strong positive selection. Thus, hybrid fitness problems are incidental byproducts of adaptive divergence, just as Darwin imagined. However, it seems that this divergence was driven not by different ecological adaptations but by genetic conflicts, involving selfish transposons. Phylogenetic tree of Hmr. The number shown above each lineage is the ratio of non-synonymous (Ka) and synonymous (Ks) substitution rates (Mol. Biol. Evol. 25, 2421 -2430, 2008).

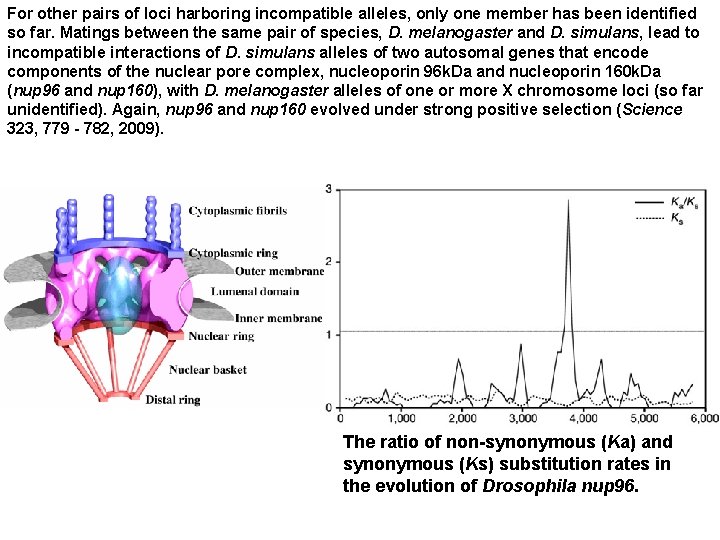
For other pairs of loci harboring incompatible alleles, only one member has been identified so far. Matings between the same pair of species, D. melanogaster and D. simulans, lead to incompatible interactions of D. simulans alleles of two autosomal genes that encode components of the nuclear pore complex, nucleoporin 96 k. Da and nucleoporin 160 k. Da (nup 96 and nup 160), with D. melanogaster alleles of one or more X chromosome loci (so far unidentified). Again, nup 96 and nup 160 evolved under strong positive selection (Science 323, 779 - 782, 2009). The ratio of non-synonymous (Ka) and synonymous (Ks) substitution rates in the evolution of Drosophila nup 96.

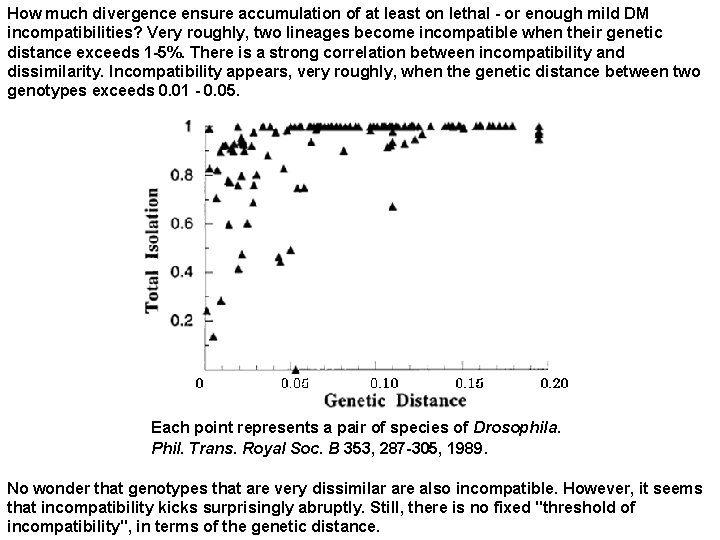
How much divergence ensure accumulation of at least on lethal - or enough mild DM incompatibilities? Very roughly, two lineages become incompatible when their genetic distance exceeds 1 -5%. There is a strong correlation between incompatibility and dissimilarity. Incompatibility appears, very roughly, when the genetic distance between two genotypes exceeds 0. 01 - 0. 05. Each point represents a pair of species of Drosophila. Phil. Trans. Royal Soc. B 353, 287 -305, 1989. No wonder that genotypes that are very dissimilar are also incompatible. However, it seems that incompatibility kicks surprisingly abruptly. Still, there is no fixed "threshold of incompatibility", in terms of the genetic distance.

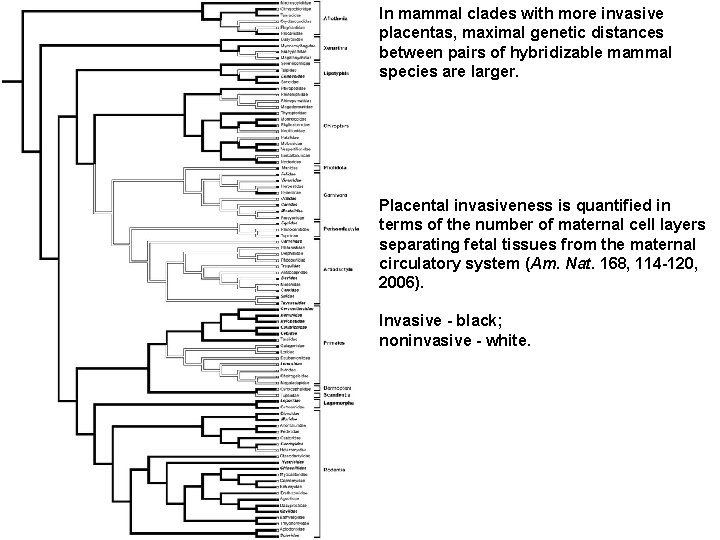
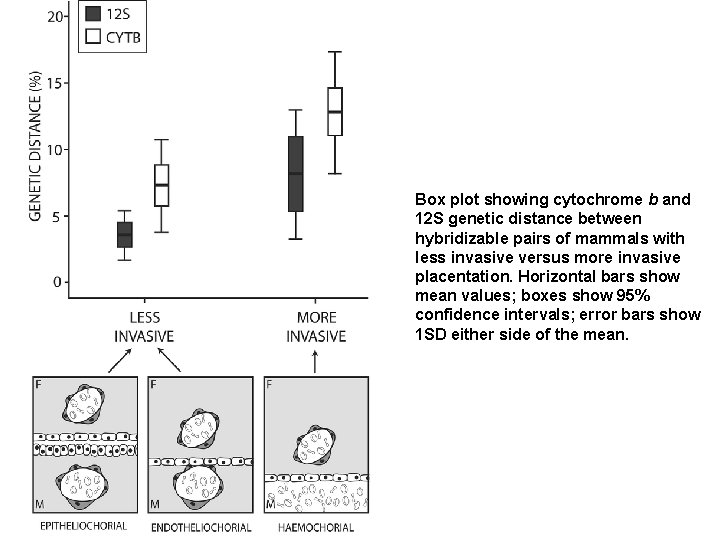
In mammal clades with more invasive placentas, maximal genetic distances between pairs of hybridizable mammal species are larger. Placental invasiveness is quantified in terms of the number of maternal cell layers separating fetal tissues from the maternal circulatory system (Am. Nat. 168, 114 -120, 2006). Invasive - black; noninvasive - white.

Box plot showing cytochrome b and 12 S genetic distance between hybridizable pairs of mammals with less invasive versus more invasive placentation. Horizontal bars show mean values; boxes show 95% confidence intervals; error bars show 1 SD either side of the mean.

Speciation, the origin of species, in a sense, origin of incompatibility between organisms. There are several modes of speciation: 1) Phyletic - in the course of evolution of one lineage it changes so profoundly that current organisms and their remote ancestors must be attributed to different species (here Zeno's paradox is obvious). Naturally, phyletic speciation is hard to study. 2) Allopatric - two lineages evolve independently, because their ranges do not overlap, and eventually become different species. Allopatric speciation is not a specific process, but just a by-product of independent divergence. 3) Sympatric - a (sexual) population splits into two species without geographic isolation. Sympatric speciation is a complex and fascinating process.

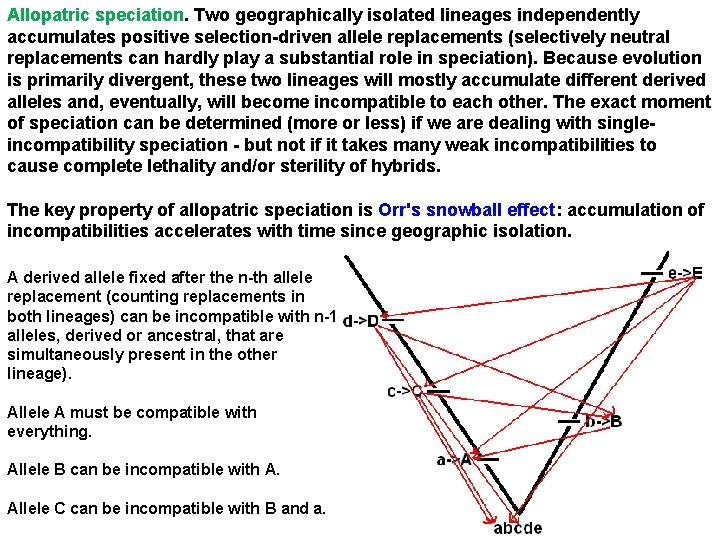
Allopatric speciation. Two geographically isolated lineages independently accumulates positive selection-driven allele replacements (selectively neutral replacements can hardly play a substantial role in speciation). Because evolution is primarily divergent, these two lineages will mostly accumulate different derived alleles and, eventually, will become incompatible to each other. The exact moment of speciation can be determined (more or less) if we are dealing with singleincompatibility speciation - but not if it takes many weak incompatibilities to cause complete lethality and/or sterility of hybrids. The key property of allopatric speciation is Orr's snowball effect: accumulation of incompatibilities accelerates with time since geographic isolation. A derived allele fixed after the n-th allele replacement (counting replacements in both lineages) can be incompatible with n-1 alleles, derived or ancestral, that are simultaneously present in the other lineage). Allele A must be compatible with everything. Allele B can be incompatible with A. Allele C can be incompatible with B and a.

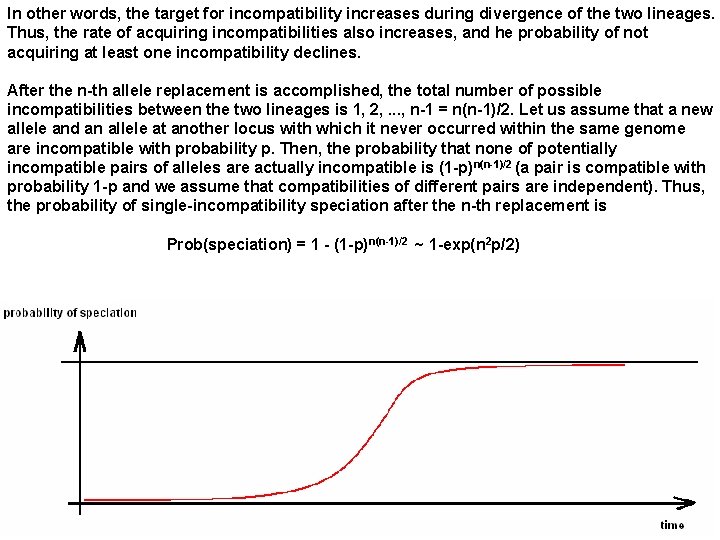
In other words, the target for incompatibility increases during divergence of the two lineages. Thus, the rate of acquiring incompatibilities also increases, and he probability of not acquiring at least one incompatibility declines. After the n-th allele replacement is accomplished, the total number of possible incompatibilities between the two lineages is 1, 2, . . . , n-1 = n(n-1)/2. Let us assume that a new allele and an allele at another locus with which it never occurred within the same genome are incompatible with probability p. Then, the probability that none of potentially incompatible pairs of alleles are actually incompatible is (1 -p) n(n-1)/2 (a pair is compatible with probability 1 -p and we assume that compatibilities of different pairs are independent). Thus, the probability of single-incompatibility speciation after the n-th replacement is Prob(speciation) = 1 - (1 -p) n(n-1)/2 ~ 1 -exp(n 2 p/2)

A physicist would say that allopatric speciation involves a phase transition - for some time, its probability is rather low, after which it rapidly reaches 100%. More realistic models assuming that speciation requires many incompatible interactions lead to similar conclusions. Allopatric speciation is a ubiquitous and unavoidable process, although the time between geographic isolation and speciation varies substantially.

Sympatric speciation of asexuals. Without sex, speciation can occur in sympatry (without spatial separation) in the same way as in allopatry, as long as ecological differentiation leads to independent regulation of densities of different organisms maintains two independently evolving sympartic lineages (such as L and S bacteria in Lenski's experiments), after which the same theory applies. Indeed, an advantageous mutation within an L individual may eventually displace all L individuals - but NOT S individuals (and vice versa). Of course, to actually measure incompatibility of asexuals is very difficult - sex is a great experimental tool for studying speciation.

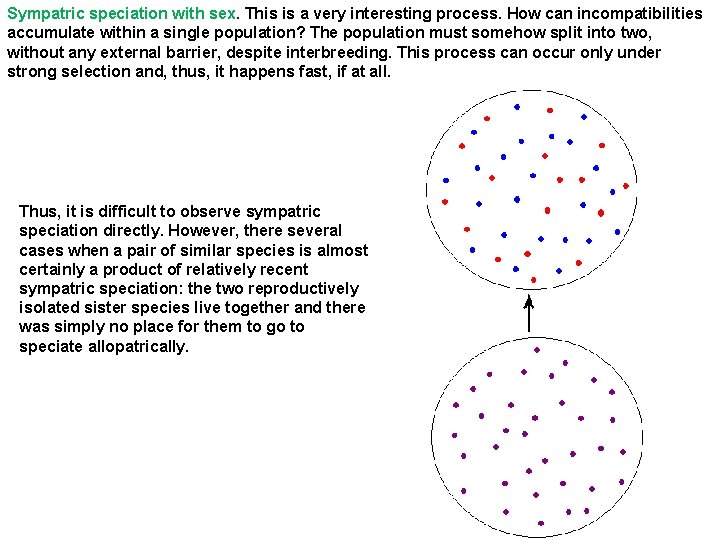
Sympatric speciation with sex. This is a very interesting process. How can incompatibilities accumulate within a single population? The population must somehow split into two, without any external barrier, despite interbreeding. This process can occur only under strong selection and, thus, it happens fast, if at all. Thus, it is difficult to observe sympatric speciation directly. However, there several cases when a pair of similar species is almost certainly a product of relatively recent sympatric speciation: the two reproductively isolated sister species live together and there was simply no place for them to go to speciate allopatrically.

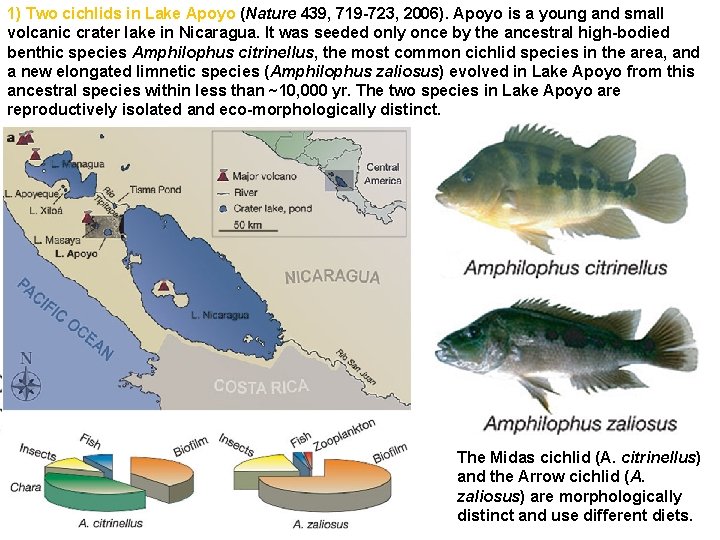
1) Two cichlids in Lake Apoyo (Nature 439, 719 -723, 2006). Apoyo is a young and small volcanic crater lake in Nicaragua. It was seeded only once by the ancestral high-bodied benthic species Amphilophus citrinellus, the most common cichlid species in the area, and a new elongated limnetic species (Amphilophus zaliosus) evolved in Lake Apoyo from this ancestral species within less than ~10, 000 yr. The two species in Lake Apoyo are reproductively isolated and eco-morphologically distinct. The Midas cichlid (A. citrinellus) and the Arrow cichlid (A. zaliosus) are morphologically distinct and use different diets.

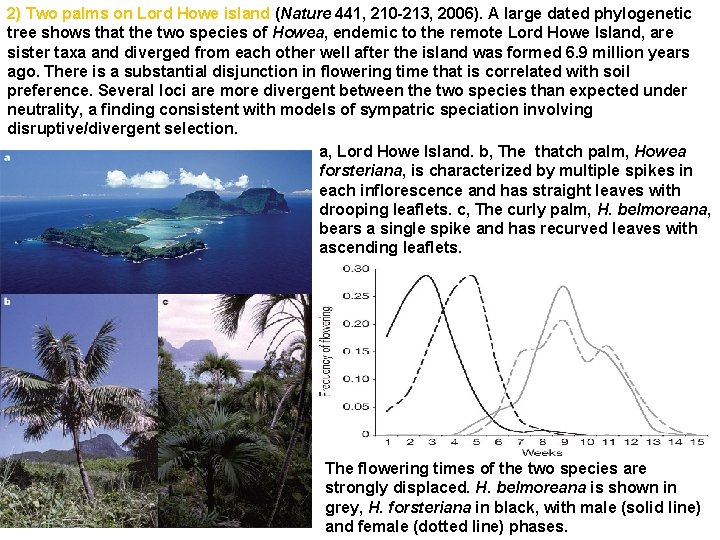
2) Two palms on Lord Howe island (Nature 441, 210 -213, 2006). A large dated phylogenetic tree shows that the two species of Howea, endemic to the remote Lord Howe Island, are sister taxa and diverged from each other well after the island was formed 6. 9 million years ago. There is a substantial disjunction in flowering time that is correlated with soil preference. Several loci are more divergent between the two species than expected under neutrality, a finding consistent with models of sympatric speciation involving disruptive/divergent selection. a, Lord Howe Island. b, The thatch palm, Howea forsteriana, is characterized by multiple spikes in each inflorescence and has straight leaves with drooping leaflets. c, The curly palm, H. belmoreana, bears a single spike and has recurved leaves with ascending leaflets. The flowering times of the two species are strongly displaced. H. belmoreana is shown in grey, H. forsteriana in black, with male (solid line) and female (dotted line) phases.

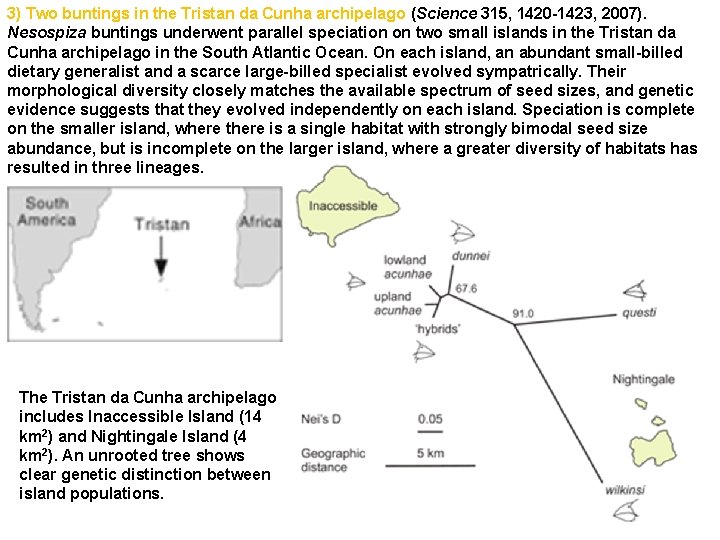
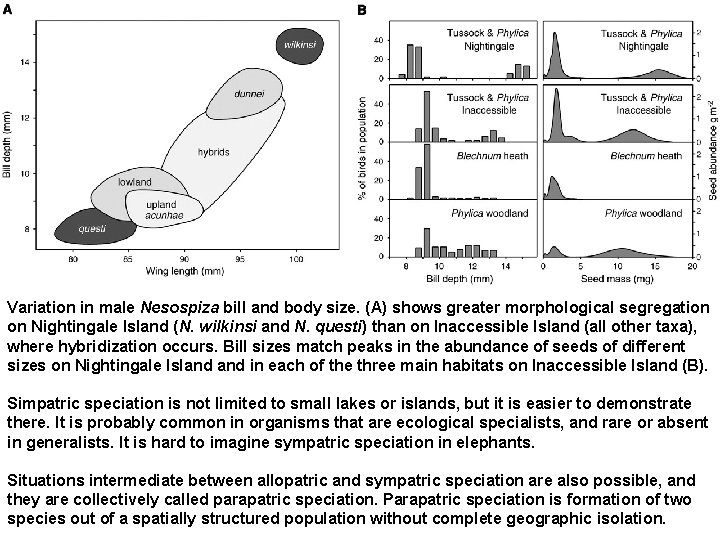
3) Two buntings in the Tristan da Cunha archipelago (Science 315, 1420 -1423, 2007). Nesospiza buntings underwent parallel speciation on two small islands in the Tristan da Cunha archipelago in the South Atlantic Ocean. On each island, an abundant small-billed dietary generalist and a scarce large-billed specialist evolved sympatrically. Their morphological diversity closely matches the available spectrum of seed sizes, and genetic evidence suggests that they evolved independently on each island. Speciation is complete on the smaller island, where there is a single habitat with strongly bimodal seed size abundance, but is incomplete on the larger island, where a greater diversity of habitats has resulted in three lineages. The Tristan da Cunha archipelago includes Inaccessible Island (14 km 2) and Nightingale Island (4 km 2). An unrooted tree shows clear genetic distinction between island populations.

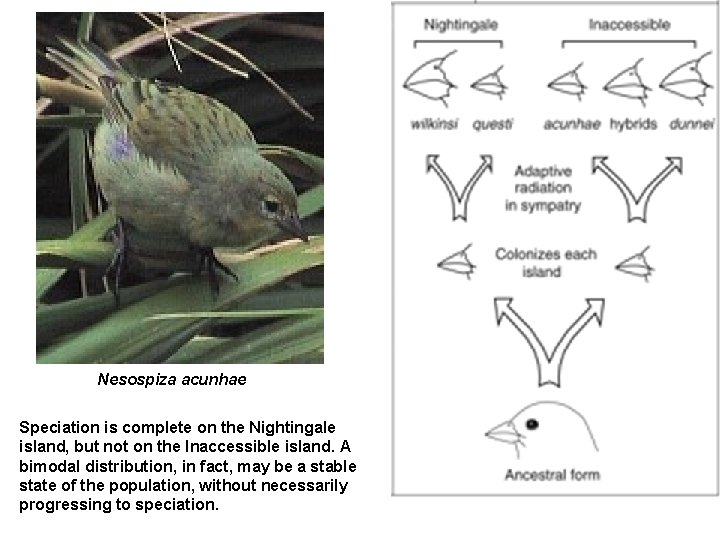
Nesospiza acunhae Speciation is complete on the Nightingale island, but not on the Inaccessible island. A bimodal distribution, in fact, may be a stable state of the population, without necessarily progressing to speciation.

Variation in male Nesospiza bill and body size. (A) shows greater morphological segregation on Nightingale Island (N. wilkinsi and N. questi) than on Inaccessible Island (all other taxa), where hybridization occurs. Bill sizes match peaks in the abundance of seeds of different sizes on Nightingale Island in each of the three main habitats on Inaccessible Island (B). Simpatric speciation is not limited to small lakes or islands, but it is easier to demonstrate there. It is probably common in organisms that are ecological specialists, and rare or absent in generalists. It is hard to imagine sympatric speciation in elephants. Situations intermediate between allopatric and sympatric speciation are also possible, and they are collectively called parapatric speciation. Parapatric speciation is formation of two species out of a spatially structured population without complete geographic isolation.

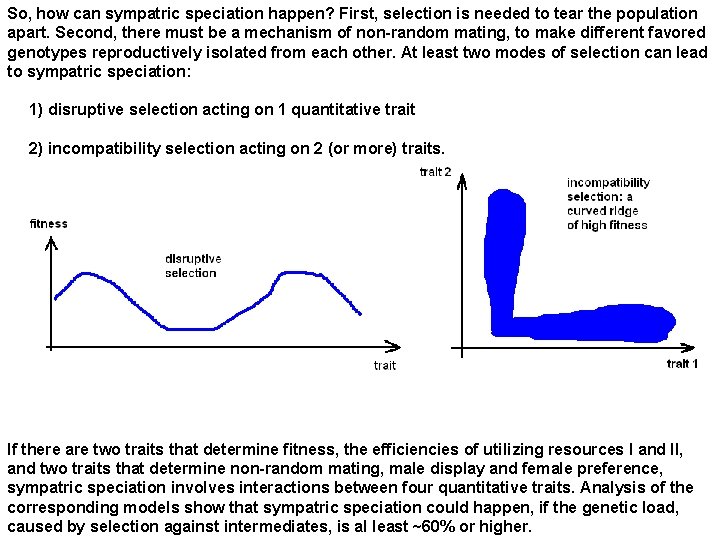
So, how can sympatric speciation happen? First, selection is needed to tear the population apart. Second, there must be a mechanism of non-random mating, to make different favored genotypes reproductively isolated from each other. At least two modes of selection can lead to sympatric speciation: 1) disruptive selection acting on 1 quantitative trait 2) incompatibility selection acting on 2 (or more) traits. If there are two traits that determine fitness, the efficiencies of utilizing resources I and II, and two traits that determine non-random mating, male display and female preference, sympatric speciation involves interactions between four quantitative traits. Analysis of the corresponding models show that sympatric speciation could happen, if the genetic load, caused by selection against intermediates, is al least ~60% or higher.

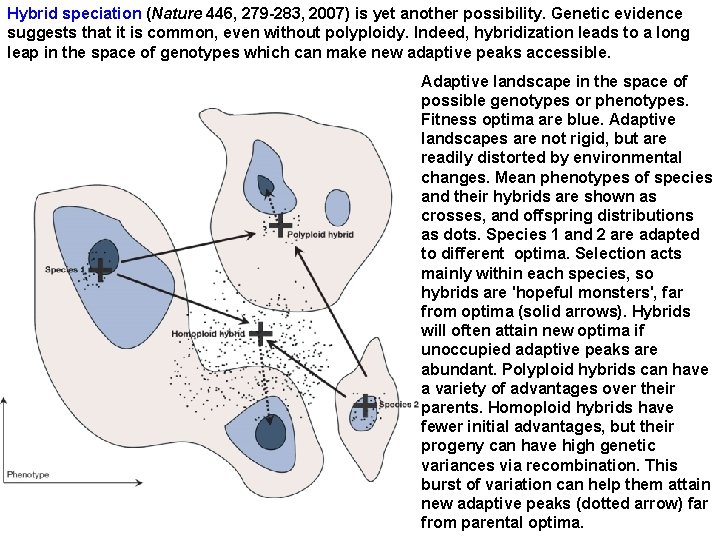
Hybrid speciation (Nature 446, 279 -283, 2007) is yet another possibility. Genetic evidence suggests that it is common, even without polyploidy. Indeed, hybridization leads to a long leap in the space of genotypes which can make new adaptive peaks accessible. Adaptive landscape in the space of possible genotypes or phenotypes. Fitness optima are blue. Adaptive landscapes are not rigid, but are readily distorted by environmental changes. Mean phenotypes of species and their hybrids are shown as crosses, and offspring distributions as dots. Species 1 and 2 are adapted to different optima. Selection acts mainly within each species, so hybrids are 'hopeful monsters', far from optima (solid arrows). Hybrids will often attain new optima if unoccupied adaptive peaks are abundant. Polyploid hybrids can have a variety of advantages over their parents. Homoploid hybrids have fewer initial advantages, but their progeny can have high genetic variances via recombination. This burst of variation can help them attain new adaptive peaks (dotted arrow) far from parental optima.

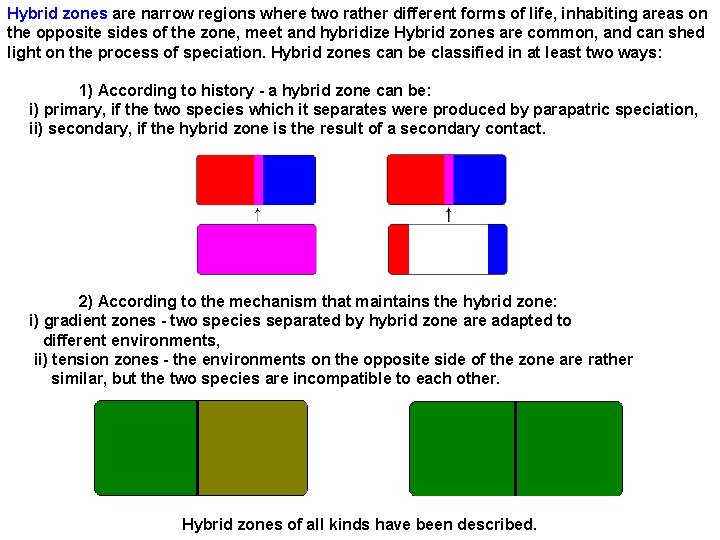
Hybrid zones are narrow regions where two rather different forms of life, inhabiting areas on the opposite sides of the zone, meet and hybridize Hybrid zones are common, and can shed light on the process of speciation. Hybrid zones can be classified in at least two ways: 1) According to history - a hybrid zone can be: i) primary, if the two species which it separates were produced by parapatric speciation, ii) secondary, if the hybrid zone is the result of a secondary contact. 2) According to the mechanism that maintains the hybrid zone: i) gradient zones - two species separated by hybrid zone are adapted to different environments, ii) tension zones - the environments on the opposite side of the zone are rather similar, but the two species are incompatible to each other. Hybrid zones of all kinds have been described.

Aquilegia formosa A. pubescens A. formosa and A. pubescens form a secondary gradient hybrid zone in Sierra Nevada. Hybrids do not show any signs of intrinsically reduced fitness.

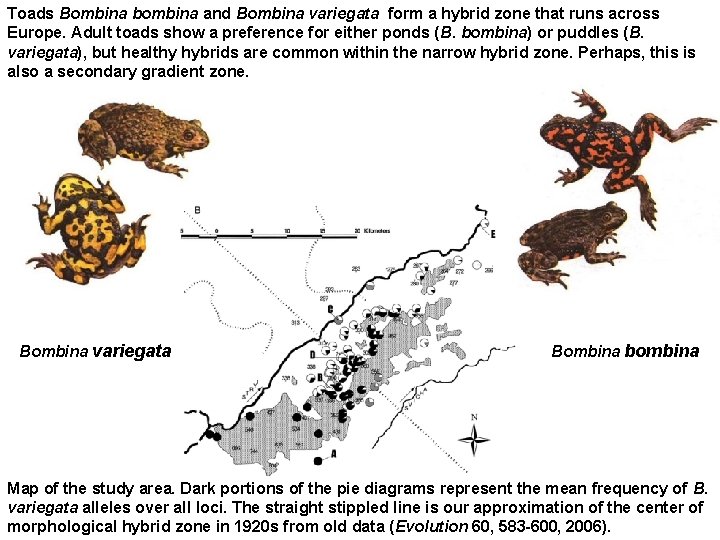
Toads Bombina bombina and Bombina variegata form a hybrid zone that runs across Europe. Adult toads show a preference for either ponds (B. bombina) or puddles (B. variegata), but healthy hybrids are common within the narrow hybrid zone. Perhaps, this is also a secondary gradient zone. Bombina variegata Bombina bombina Map of the study area. Dark portions of the pie diagrams represent the mean frequency of B. variegata alleles over all loci. The straight stippled line is our approximation of the center of morphological hybrid zone in 1920 s from old data (Evolution 60, 583 -600, 2006).

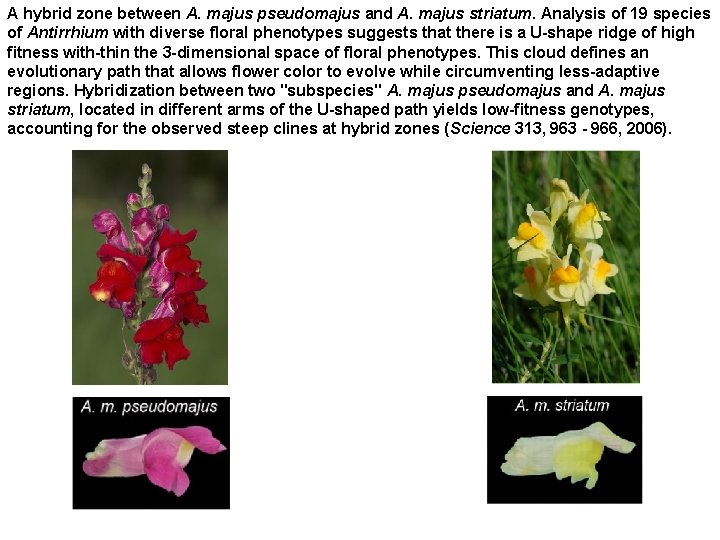
A hybrid zone between A. majus pseudomajus and A. majus striatum. Analysis of 19 species of Antirrhium with diverse floral phenotypes suggests that there is a U-shape ridge of high fitness with-thin the 3 -dimensional space of floral phenotypes. This cloud defines an evolutionary path that allows flower color to evolve while circumventing less-adaptive regions. Hybridization between two "subspecies" A. majus pseudomajus and A. majus striatum, located in different arms of the U-shaped path yields low-fitness genotypes, accounting for the observed steep clines at hybrid zones (Science 313, 963 - 966, 2006).

Flowers of 19 Antirrhinum species used for studying the space of flower phenotypes.

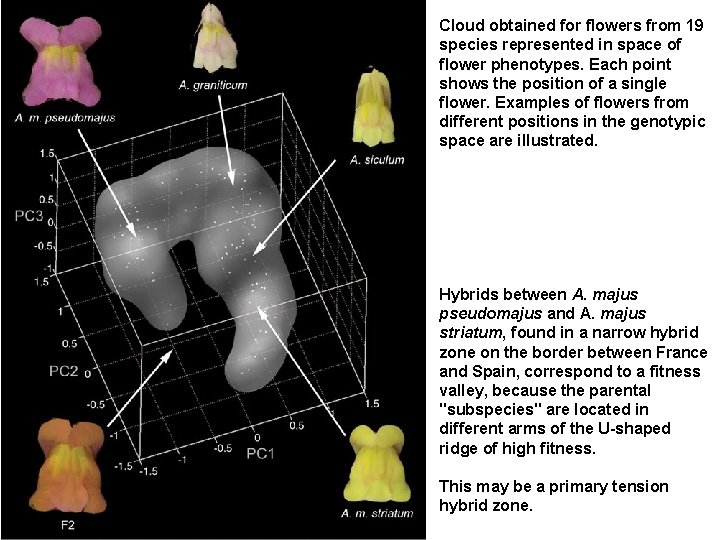
Cloud obtained for flowers from 19 species represented in space of flower phenotypes. Each point shows the position of a single flower. Examples of flowers from different positions in the genotypic space are illustrated. Hybrids between A. majus pseudomajus and A. majus striatum, found in a narrow hybrid zone on the border between France and Spain, correspond to a fitness valley, because the parental "subspecies" are located in different arms of the U-shaped ridge of high fitness. This may be a primary tension hybrid zone.

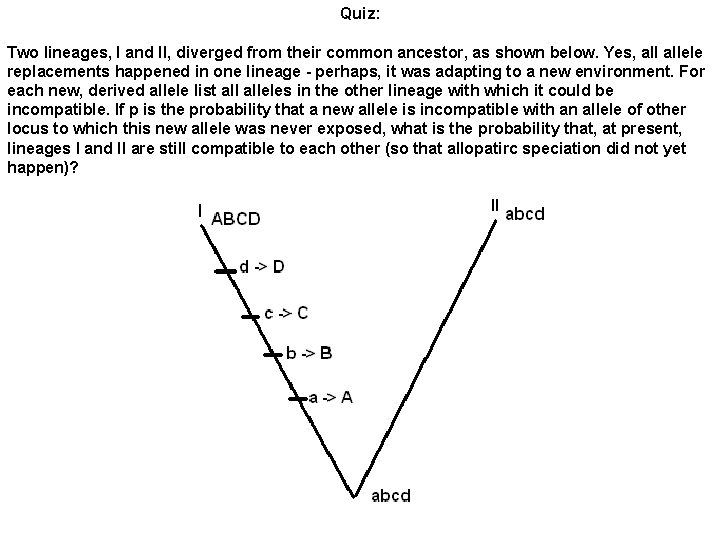
Quiz: Two lineages, I and II, diverged from their common ancestor, as shown below. Yes, allele replacements happened in one lineage - perhaps, it was adapting to a new environment. For each new, derived allele list alleles in the other lineage with which it could be incompatible. If p is the probability that a new allele is incompatible with an allele of other locus to which this new allele was never exposed, what is the probability that, at present, lineages I and II are still compatible to each other (so that allopatirc speciation did not yet happen)?