R Packages for GenomeWide Association Studies Qunyuan Zhang









- Slides: 18

R Packages for Genome-Wide Association Studies Qunyuan Zhang Division of Statistical Genomics Statistical Genetics Forum March 10, 2008

What is R ? u. R is a free software environment for statistical computing and graphics. u. Run s on a wide variety of UNIX platforms, Windows and Mac. OS (interactive or batch mode) u. Free and open source, can be downloaded from cran. r-project. org u. Wide range of packages (base & contributed), novel methods available u. Concise u. Help grammar & good structure (function, data object, methods and class) from manuals and email group u. Slow, time and memory consuming (can be overcome by parallel computation, and/or integration with C) u. Popular, used by 70~80% statisticians

R Task Views http: //cran. r-project. org/web/views/

Statistical Genetics Packages in R http: //cran. r-project. org/web/views/Genetics. html Population Genetics : genetics (basic), Geneland (spatial structures of genetic data), rmetasim (population genetics simulations), hapsim (simulation), popgen (clustering SNP genotype data and SNP simulation), hierfstat (hierarchical F-statistics of genetic data), hwde (modeling genotypic disequilibria), Biodem (biodemographical analysis), kinship (pedigree analysis), adegenet (population structure), ape & ap. Treeshape (Phylogenetic and evolution analyses), ouch (Ornstein-Uhlenbeck models), PHYLOGR (simulation and GLS model), stepwise (recombination breakpoints) Linkage and Association : gap (both population and family data, sample size calculations, probability of familial disease aggregation, kinship calculation, linkage and association analyses, haplotype frequencies) tdthap (TDT for haplotypes, powerpkg (power analyses for the affected sib pair and the TDT design), hapassoc (likelihood inference of trait associations with haplotypes in GLMs), haplo. ccs (haplotype and covariate relative risks in case-control data by weighted logistic regression), haplo. stats (haplotype analysis for unrelated subjects), tdthap (haplotype transmission/disequilibrium tests), ld. Design (experiment design for association and LD studies), LDheatmap (heatmap of pairwise LD), . map. LD (LD and haplotype blocks), pbat. R (R version of PBAT), Gen. ABEL & SNPassoc for GWAS QTL mapping for the data from experimental crosses: bqtl (inbred crosses and recombinant inbred lines), qtl (genome-wide scans), qtl. Design (designing QTL experiments & power computations), qtlbim (Bayesian Interval QTL Mapping) Sequence & Array Data Processing : seqinr, Bio. Conductor packages

Gen. ABEL Aulchenko Y. S. , Ripke S. , Isaacs A. , van Duijn C. M. Gen. ABEL: an R package for genome-wide association analysis. Bioinformatics. 2007, 23(10): 1294 -6. Gen. ABEL: genome-wide SNP association analysis a package for genome-wide association analysis between quantitative or binary traits and single-nucleotides polymorphisms (SNPs). Version: 1. 3 -5 Depends: R (≥ 2. 4. 0), methods, genetics, haplo. stats, qvalue, MASS Date: 2008 -02 -17 Author: Yurii Aulchenko, with contributions from Maksim Struchalin, Stephan Ripke and Toby Johnson Maintainer: Yurii Aulchenko <i. aoultchenko at erasmusmc. nl> License: GPL (≥ 2) In views: Genetics CRAN checks: Gen. ABEL results

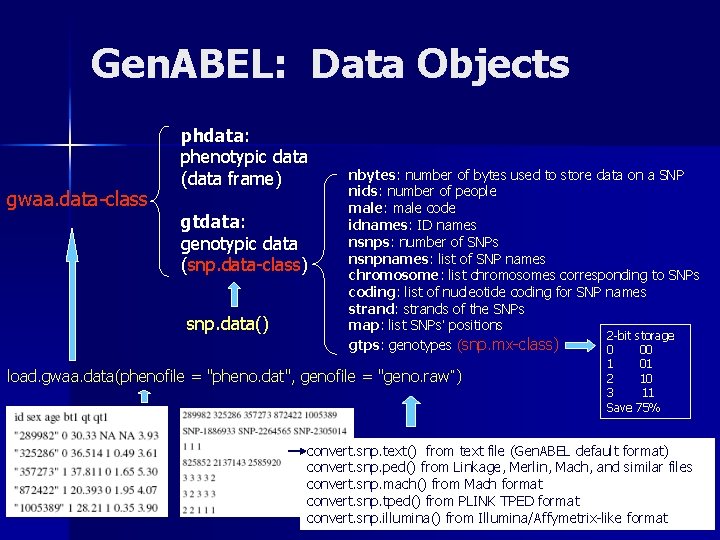
Gen. ABEL: Data Objects gwaa. data-class phdata: phenotypic data (data frame) gtdata: genotypic data (snp. data-class) snp. data() nbytes: number of bytes used to store data on a SNP nids: number of people male: male code idnames: ID names nsnps: number of SNPs nsnpnames: list of SNP names chromosome: list chromosomes corresponding to SNPs coding: list of nucleotide coding for SNP names strand: strands of the SNPs map: list SNPs’ positions 2 -bit storage gtps: genotypes (snp. mx-class) 0 00 load. gwaa. data(phenofile = "pheno. dat", genofile = "geno. raw“) 1 2 3 Save 01 10 11 75% convert. snp. text() from text file (Gen. ABEL default format) convert. snp. ped() from Linkage, Merlin, Mach, and similar files convert. snp. mach() from Mach format convert. snp. tped() from PLINK TPED format convert. snp. illumina() from Illumina/Affymetrix-like format

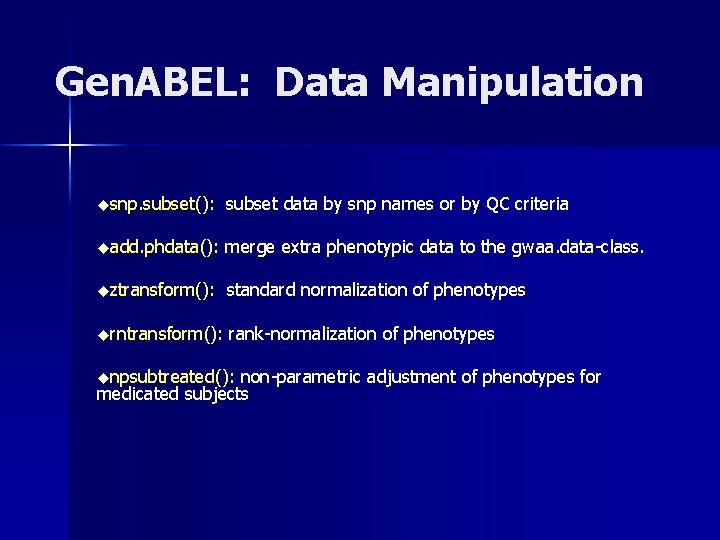
Gen. ABEL: Data Manipulation usnp. subset(): subset data by snp names or by QC criteria uadd. phdata(): merge extra phenotypic data to the gwaa. data-class. uztransform(): standard normalization of phenotypes urntransform(): rank-normalization of phenotypes unpsubtreated(): non-parametric adjustment of phenotypes for medicated subjects

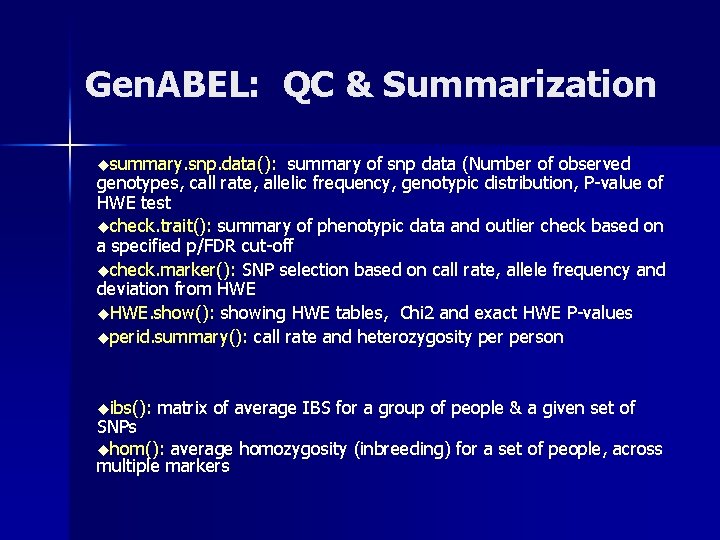
Gen. ABEL: QC & Summarization usummary. snp. data(): summary of snp data (Number of observed genotypes, call rate, allelic frequency, genotypic distribution, P-value of HWE test ucheck. trait(): summary of phenotypic data and outlier check based on a specified p/FDR cut-off ucheck. marker(): SNP selection based on call rate, allele frequency and deviation from HWE u. HWE. show(): showing HWE tables, Chi 2 and exact HWE P-values uperid. summary(): call rate and heterozygosity person uibs(): matrix of average IBS for a group of people & a given set of SNPs uhom(): average homozygosity (inbreeding) for a set of people, across multiple markers

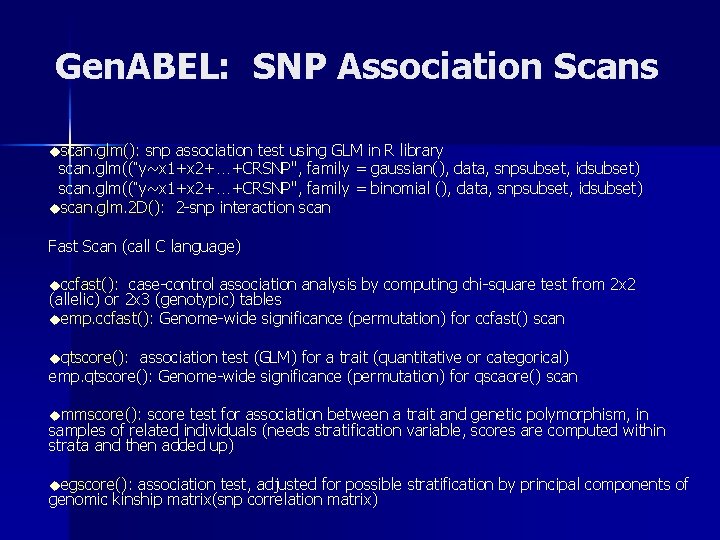
Gen. ABEL: SNP Association Scans uscan. glm(): snp association test using GLM in R library scan. glm((“y~x 1+x 2+…+CRSNP", family = gaussian(), data, snpsubset, idsubset) scan. glm((“y~x 1+x 2+…+CRSNP", family = binomial (), data, snpsubset, idsubset) uscan. glm. 2 D(): 2 -snp interaction scan Fast Scan (call C language) uccfast(): case-control association analysis by computing chi-square test from 2 x 2 (allelic) or 2 x 3 (genotypic) tables uemp. ccfast(): Genome-wide significance (permutation) for ccfast() scan uqtscore(): association test (GLM) for a trait (quantitative or categorical) emp. qtscore(): Genome-wide significance (permutation) for qscaore() scan ummscore(): score test for association between a trait and genetic polymorphism, in samples of related individuals (needs stratification variable, scores are computed within strata and then added up) uegscore(): association test, adjusted for possible stratification by principal components of genomic kinship matrix(snp correlation matrix)

Gen. ABEL: Haplotype Association Scans uscan. haplo(): haplotype association test using GLM in R library uscan. haplo. 2 D(): 2 -haplotype interaction scan (haplo. stats package required) Sliding window strategy Posterior prob. of Haplotypes via EM algorithm GLM-based score test for haplotype-trait association (Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. 2002. Score tests for association of traits with haplotypes when linkage phase is ambiguous Am J Hum Genet 70: 425 -434. )

Gen. ABEL: GWAS results from scan. glm, scan. haplo, ccfast, qtscore, emp. ccfast, emp. qtscore scan. gwaa-class u. Names: snpnames list of names of SNPs tested u. P 1 df: p-values of 1 -d. f. (additive or allelic) test for association u. P 2 df: p-values of 2 -d. f. (genotypic) test for association u. Pc 1 df: p-values from the 1 -d. f. test for association between SNP and trait; the statistics is corrected for possible inflation ueff. B: effect of the B allele in allelic test ueff. AB: effect of the AB genotype in genotypic test ueff. BB: effect of the BB genotype in genotypic test u. Map: list of map positions of the SNPs u. Chromosome: list of chromosomes the SNPs belong to u. Idnames: list of subjects used in analysis u. Lambda: inflation factor estimate, as computed using lower portion (say, 90%) of the distribution, and standard error of the estimate u. Formula: formula/function used to compute p-values u. Family: family of the link function / nature of the test

Gen. ABEL: Table & Graphic Functions descriptives. marker(): descriptives. trait(): descriptives. scan(): table of marker info. table of trait info. table of scan results plot. scan. gwaa(): plot of scan results plot. check. marker(): plot of marker data (QC etc. )

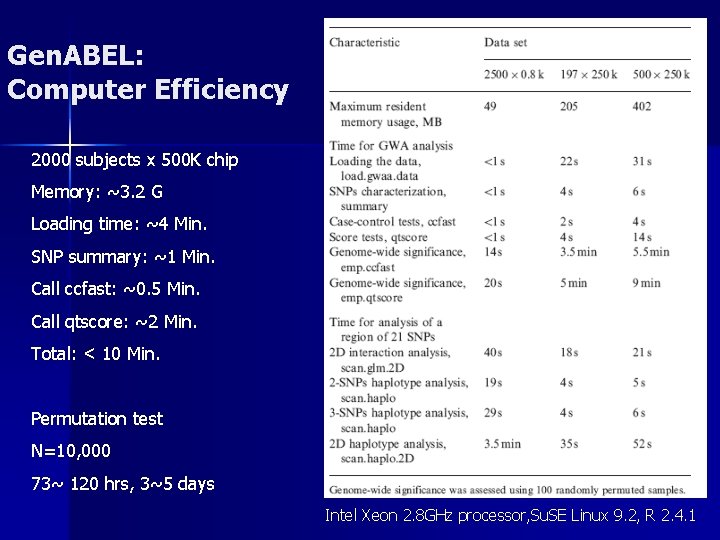
Gen. ABEL: Computer Efficiency 2000 subjects x 500 K chip Memory: ~3. 2 G Loading time: ~4 Min. SNP summary: ~1 Min. Call ccfast: ~0. 5 Min. Call qtscore: ~2 Min. Total: < 10 Min. Permutation test N=10, 000 73~ 120 hrs, 3~5 days Intel Xeon 2. 8 GHz processor, Su. SE Linux 9. 2, R 2. 4. 1

SNPassoc An R package to perform whole genome association studies, Juan R. González 1, et al. Bioinformatics, 2007 23(5): 654 -655 SNPassoc: SNPs-based whole genome association studies This package carries out most common analysis when performing whole genome association studies. These analyses include descriptive statistics and exploratory analysis of missing values, calculation of Hardy-Weinberg equilibrium, analysis of association based on generalized linear models (either for quantitative or binary traits), and analysis of multiple SNPs (haplotype and epistasis analysis). Permutation test and related tests (sum statistic and truncated product) are also implemented. Version: 1. 4 -9 Depends: R (≥ 2. 4. 0), haplo. stats, survival, mvtnorm Date: 2007 -Oct-16 Author: Juan R González, Lluís Armengol, Elisabet Guinó, Xavier Solé, and Víctor Moreno. Maintainer: Juan R González <jrgonzalez at imim. es> License: GPL version 2 or newer. URL: http: //www. r-project. org and http: //davinci. crg. es/estivill_lab/snpassoc; In views: Genetics CRAN checks: SNPassoc results

SNPassoc: Data & Summary usetup. SNP(data=snp-pheno. table, col. SNPs=, sep = "/", . . . ) usummary() allele frequencies percentage of missing values HWE test info=map. table,

SNPassoc: Association Tests u. WGassociation(y~x 1+x 2, data=, model = (codominant, recessive, overdominant, log-additive or all), quantitative = , level = 0. 95) uscan. WGassociation(): only p values uassociation(): only for selected snps, can do stratified, Gx. E interaction analyses Results ØSummary: a summary table by genes/chromosomes ØWgstats: detailed output(case-control numbers, percentages, odds ratios/ mean differences, 95% confidence intervals, P-value for the likelihood ratio test of association, and AIC, etc. ) ØPvalues: a table of p-values for each genetic model for each SNP ØPlot: p values in the -log scale for plot. Wgassociation() ØLabels: returns the names of the SNPs analyzed

SNPassoc: Multiple-SNP Analysis SNP–SNP Interaction interaction. Pval(): epistasis analysis between all pairs of SNPs (and covariates). Haplotype Analysis haplo. glm(): using the R package haplo. stats: association analysis of haplotypes with a response via GLM haplo. interaction(): interactions between haplotypes (and covariates)

SNPassoc: Computer Efficiency 1000 subjects X 3000 SNPs 5 min. import data 40 min. setup. SNP() 30 min. scan. WGassociation(): only p values (including permutation test) Memory usage: 750 MB
 Paradigm shift from women studies to gender studies
Paradigm shift from women studies to gender studies Modern studies association
Modern studies association Smd ic packages
Smd ic packages Ssisunit
Ssisunit Zscaler packages
Zscaler packages Microfilament function
Microfilament function Mtc prepaid
Mtc prepaid Dts sql server 2005
Dts sql server 2005 Big things come in small packages eleanora e tate
Big things come in small packages eleanora e tate Ecommerce packages 2
Ecommerce packages 2 London rangers youth and community
London rangers youth and community Bsd packages
Bsd packages All crm packages contain modules for prm and erm.
All crm packages contain modules for prm and erm. Access quarterly package
Access quarterly package Le meridien cairo airport wedding packages
Le meridien cairo airport wedding packages Connectivity package abb
Connectivity package abb Unified product and services
Unified product and services One week carlos bought 2 packages of dog bones
One week carlos bought 2 packages of dog bones Sublime text package control install
Sublime text package control install