Protein dynamics n Foldingunfolding dynamics Passage over one








- Slides: 18

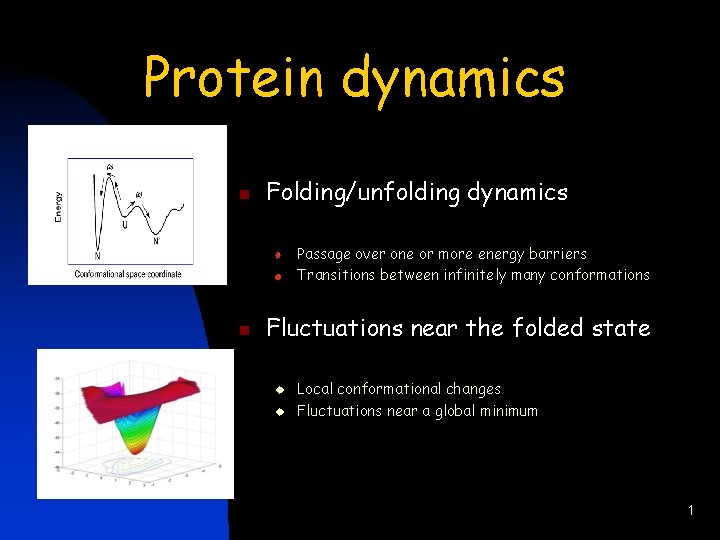
Protein dynamics n Folding/unfolding dynamics Passage over one or more energy barriers Transitions between infinitely many conformations n Fluctuations near the folded state u u Local conformational changes Fluctuations near a global minimum 1

Fluctuations near folded structure 2

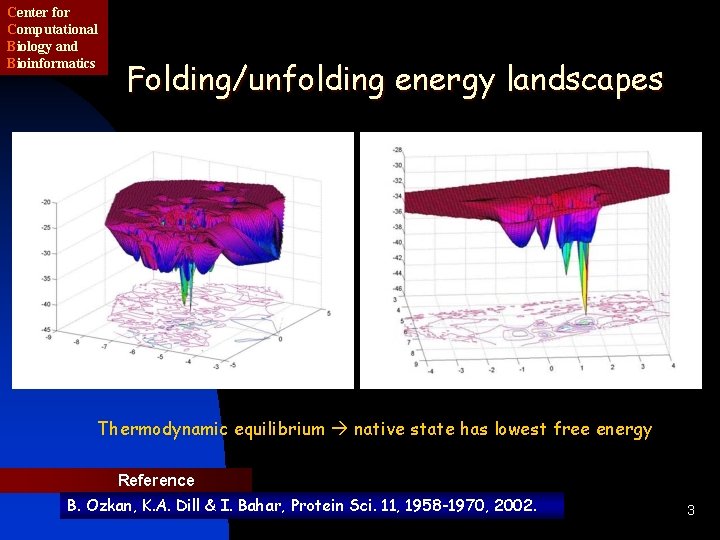
Center for Computational Biology and Bioinformatics Folding/unfolding energy landscapes Thermodynamic equilibrium native state has lowest free energy Reference B. Ozkan, K. A. Dill & I. Bahar, Protein Sci. 11, 1958 -1970, 2002. 3

Knowledge-based studies Exploiting PDB structures. . .

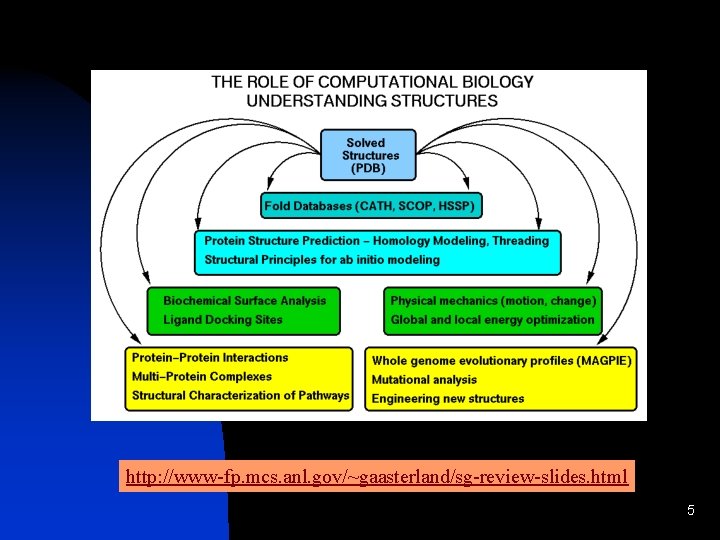
http: //www-fp. mcs. anl. gov/~gaasterland/sg-review-slides. html 5

Protein folding problem: “Predicting 3 -dimensional structure from amino acid sequence” n n n A unique folded structure (native conformation, native fold) is assumed by a given sequence, although infinitely many conformations can be accessed. Which? (Protein folding problem) How, why? (Folding kinetics) Basic postulate: Thermodynamic equilibrium Global energy minimum CASP (Critical Assessment of Structure Prediction) 6

Protein structure prediction Three computational methods: • Homology modeling • Threading • Ab initio simulations 9/25/2020 7

Homology/comparative modeling MODELLER is used for homology or comparative modeling of protein three-dimensional structures (1). The user provides an alignment of a sequence to be modeled with known related structures and MODELLER automatically calculates a model containing all non-hydrogen atoms. MODELLER implements comparative protein structure modeling by satisfaction of spatial restraints (2, 3), and can perform many additional tasks, including de novo modeling of loops in protein structures, optimization of various models of protein structure with respect to a flexibly defined objective function, multiple alignment of protein sequences and/or structures, clustering, searching of sequence databases, comparison of protein structures, etc. MODELLER is written in Fortran 90 and runs on the Pentium PC's (Linux and Win XP), Apple Macintosh (OS X) and workstations from Silicon Graphics (IRIX), Sun (Solaris), IBM (AIX), and DEC Alpha (OSF/1). http: //guitar. rockefeller. edu/modeller. html (A. Sali) 8

SWISS-MODEL An Automated Comparative Protein Modelling Server accessible via the Ex. PASy (Expert Protein Analysis System) web server (by Peitsch et al. ) STEPS: 1. Search for suitable templates (from Ex. NRL-3 D , using BLAST) 2. Check sequence identity with target (SIM will select all templates with sequence identities above 25% and N> 20) 3. Create Pro. Mod. II jobs 4. Generate models (Pro. Mod. II) using known 3 -d templates 5. Energy minimization with Gromos 96 http: //swissmodel. expasy. org/SWISS-MODEL. html 9

Predict Protein (Rost) http: //www. embl-heidelberg. de/predictprotein/ 10

Structural Homology n Dali Server (Sander-Holm) http: //www 2. ebi. ac. uk/dali/ L. Holm and C. Sander (1996) Mapping the protein universe. Science 273: 595 -602. 11

Threading (Fold recognition) Loopp (Elber) Threader (Jones) 12

Ab initio simulations n n n Protarch (Scheraga’s group) Rosetta (Baker’s lab) Touchstone (Skolnick) 13

Need for Low Resolution Approaches Coarse-grained Models with Empirical Force Fields are the most tractable - if not the only possible – computational tools for investigating large systems, and complex biological processes 14

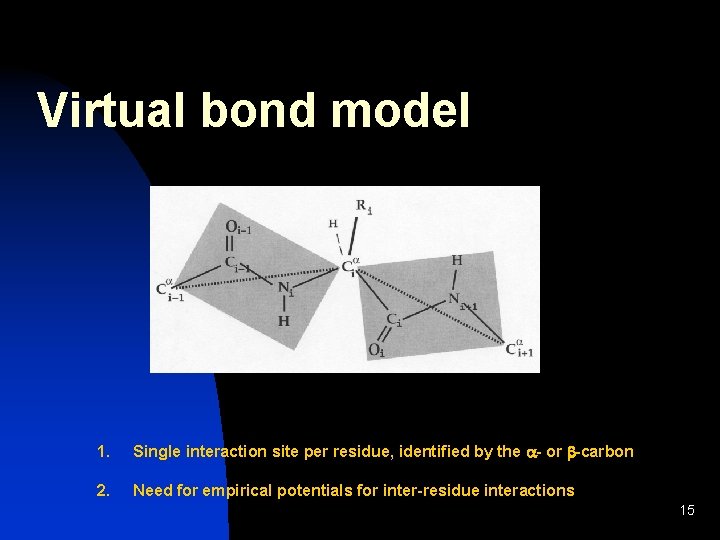
Virtual bond model 1. Single interaction site per residue, identified by the a- or b-carbon 2. Need for empirical potentials for inter-residue interactions 15

Swiss-Pdb. Viewer Free, powerful and easy to use Program Download: http: //us. expasy. org/spdbv/mainpage. htm Manual Download: http: //us. expasy. org/spdbv/program/Deep. View. Manual. pdf Sample Files Download: 1 A 30, 1 A 8 G from PDB bank: http: //www. rcsb. org/pdb/ 9/25/2020 16

Tutorial pages: http: //us. expasy. org/spdbv/text/tutorial. htm http: //www. usm. maine. edu/%7 Erhodes/SPVTut/index. html 1. Load a file & pop up functional Windows and panels Make sure you activate control panel, alignment windows every time you load a file 2. Selection, Display, Color Practice: a. Mark all the lysine in chain A red in Ribbon diagram b. Color all the alpha-helix by their solvent accessibility c. Color all the non-polar residues by their Temp-B factor 3. Handy icons & move all or move selection Common used functions: measure atom-atom distance; mutate a residue Use Esc when you want to jump out a certain function! 17

4. Computing tools H-bond Surface Energy minimization 5. Structure alignment (Fit) Switch between active layers Visible check box, move check box 6. Homology Modeling (SWISS MODEL) 7. Output – Save as Fig, Save the modified layer 8. Script commands & running script Have fun. Play around with it! Contact: Lee lwy 1@pitt. edu at CCBB 18