Oxford Nanopore Technologies Nanopore Sequencing Introduction to nanopore






- Slides: 17

Oxford Nanopore Technologies Nanopore Sequencing

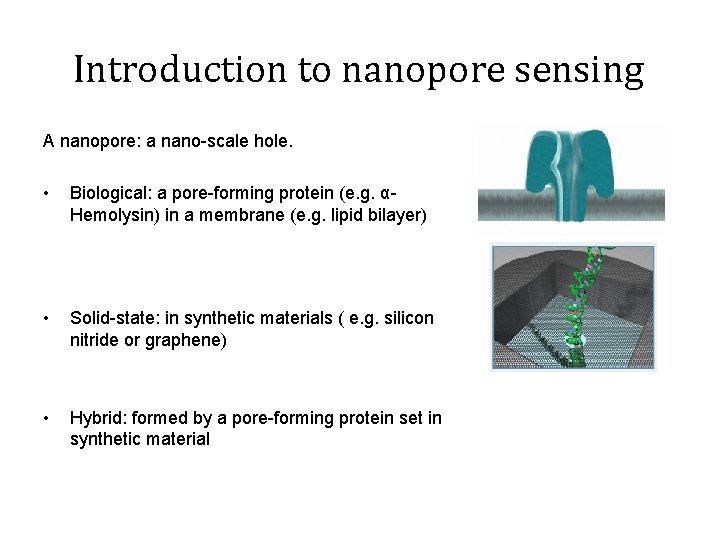
Introduction to nanopore sensing A nanopore: a nano-scale hole. • Biological: a pore-forming protein (e. g. αHemolysin) in a membrane (e. g. lipid bilayer) • Solid-state: in synthetic materials ( e. g. silicon nitride or graphene) • Hybrid: formed by a pore-forming protein set in synthetic material

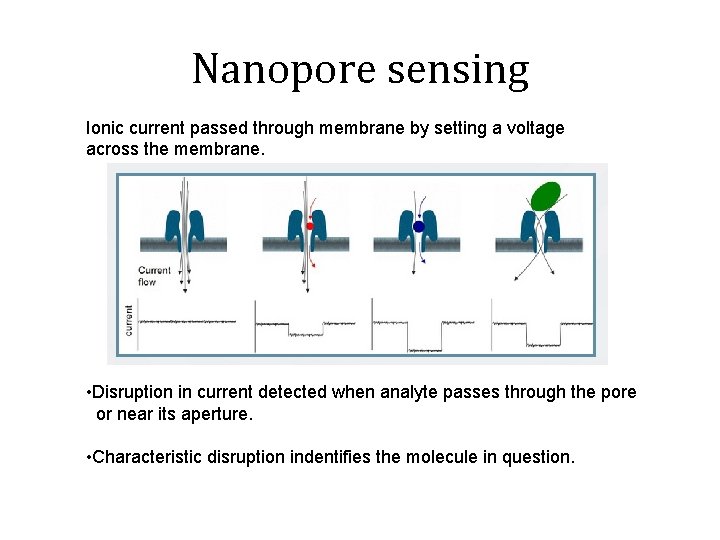
Nanopore sensing Ionic current passed through membrane by setting a voltage across the membrane. • Disruption in current detected when analyte passes through the pore or near its aperture. • Characteristic disruption indentifies the molecule in question.

Nanopore DNA sequencing • DNA polymer or individual nucleotides pass through the nanopore. • Detected by – a adaptor molecule ( e. g. Cyclodextrin). – Tunnelling electrodes based detectors. – Capacitive detectors – Graphene based nano-gap or edge state detectors.

Nanopore DNA sequencing • Strand sequencing: – Sequencing in real-time as the intact DNA polymer passes through the nanopore. • Exonuclease sequencing: – Individual nucleotides pass through the nanopore by the aid of processive exonuclease.

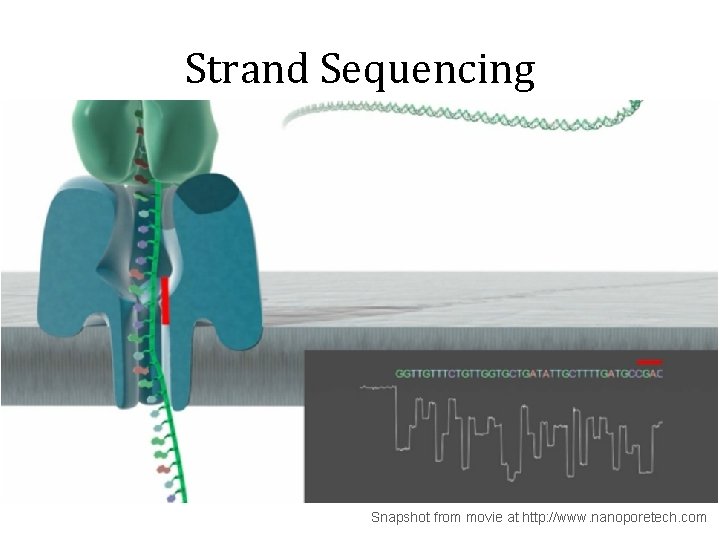
Strand Sequencing Snapshot from movie at http: //www. nanoporetech. com

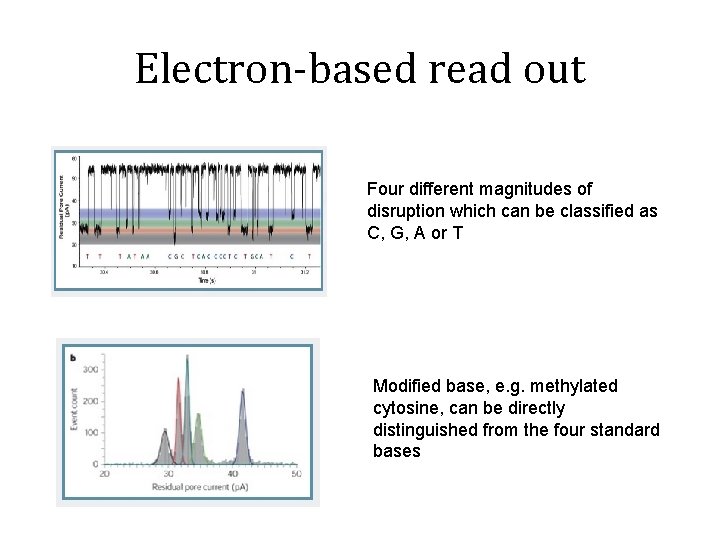
Electron-based read out Four different magnitudes of disruption which can be classified as C, G, A or T Modified base, e. g. methylated cytosine, can be directly distinguished from the four standard bases

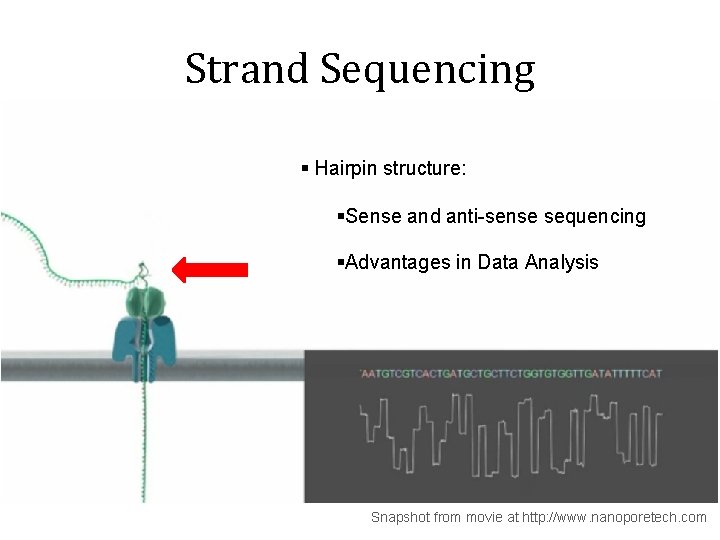
Strand Sequencing § Hairpin structure: §Sense and anti-sense sequencing §Advantages in Data Analysis Snapshot from movie at http: //www. nanoporetech. com

Exonuclease Sequencing Snapshot from movie at http: //www. nanoporetech. com

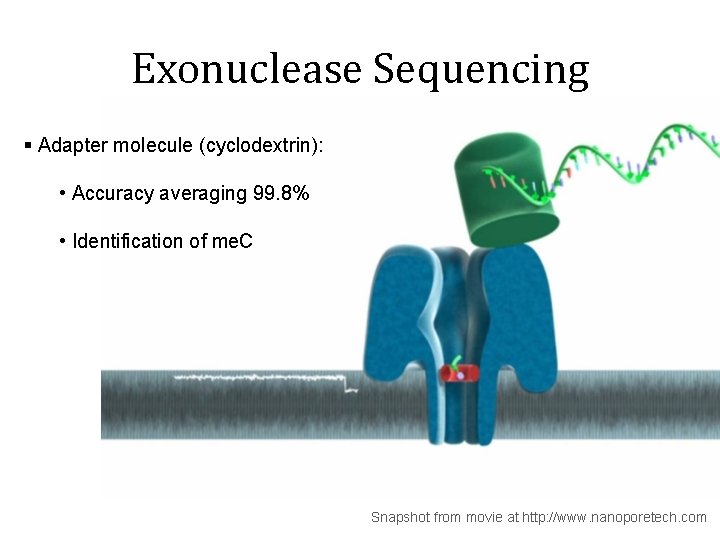
Exonuclease Sequencing § Adapter molecule (cyclodextrin): • Accuracy averaging 99. 8% • Identification of me. C Snapshot from movie at http: //www. nanoporetech. com

Working strategy • Min. ION: a miniaturised sensing instrument – Portable. – Field-deployable. – Requires minimal sample prep. – Compatible with blood serum, plasma and whole blood.

Working strategy • Grid. ION system – Uses single-use, self-contained cartridge. – Can be used as a single instrument: Node – Can be used in a cluster, connected through network. – Low power and space required. – Permits scheduling and multiplexing.

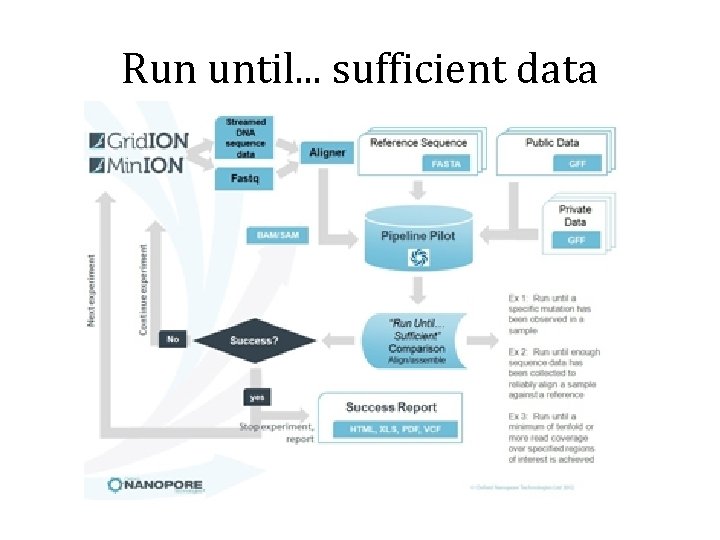
Workflow versatility • No fixed run time – Can be run one or more nodes for minutes or days. – Data analysis takes place in real time. – Longer run enables collecting more data points. • Run until. . . sufficient data – The Grid. ION system enables users to run an experiment until sufficient data has been collected to reach a predetermined experimental endpoint.

Run until. . . sufficient data

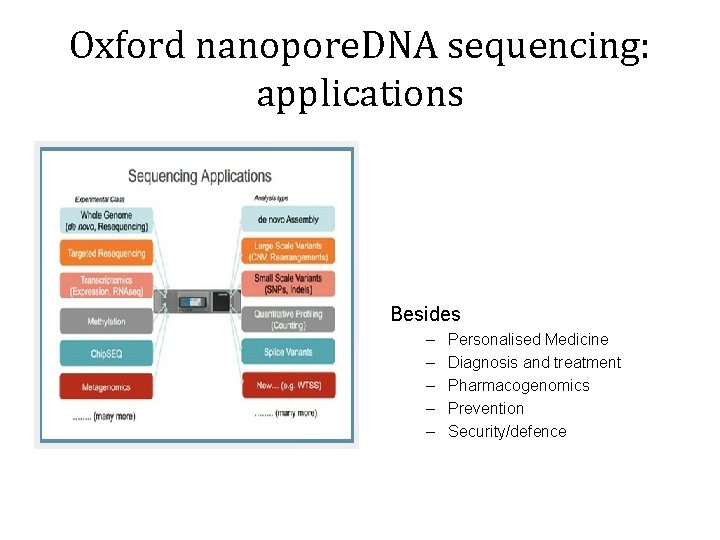
Oxford nanopore. DNA sequencing: applications Besides – – – Personalised Medicine Diagnosis and treatment Pharmacogenomics Prevention Security/defence

Advantages over present sequencing technologies • • Real-time sequencing strategy. No strand amplification needed. No bias due to sequencing amplification. Low cost: trying to fulfil the target of $1000 per human genome. Lager read size: read size is limited only by preparation. No requirement for large amounts of high-performance disk storage. Large-scale structural variation can be detected at lower depth of coverage. • Enable long-range haplotyping. • No need for expensive and time-consuming mate pair library construction.

Thank you
 Oxford nanopore
Oxford nanopore Bongcam-rudloff
Bongcam-rudloff Tet d
Tet d Introduction to wan technologies
Introduction to wan technologies Emerging technologies introduction
Emerging technologies introduction Ap csp sequencing
Ap csp sequencing Exome sequencing project
Exome sequencing project Sequence expository text examples
Sequence expository text examples Check valve symbol
Check valve symbol In which things
In which things Cyclical scheduling example
Cyclical scheduling example Rna sequencing steps
Rna sequencing steps What is sequencing selection and iteration
What is sequencing selection and iteration Microprogram sequencer block diagram
Microprogram sequencer block diagram Johnson's rule of sequencing
Johnson's rule of sequencing Job sequence
Job sequence Illumina sequencing chemistry
Illumina sequencing chemistry Cask of amontillado quiz
Cask of amontillado quiz