Origins of life MillerUrey experiment 1953 A M












- Slides: 32

Origins of life



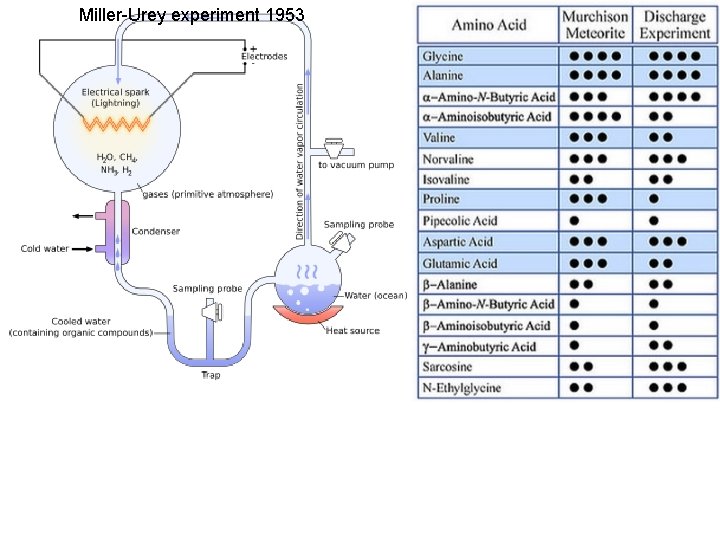
Miller-Urey experiment 1953



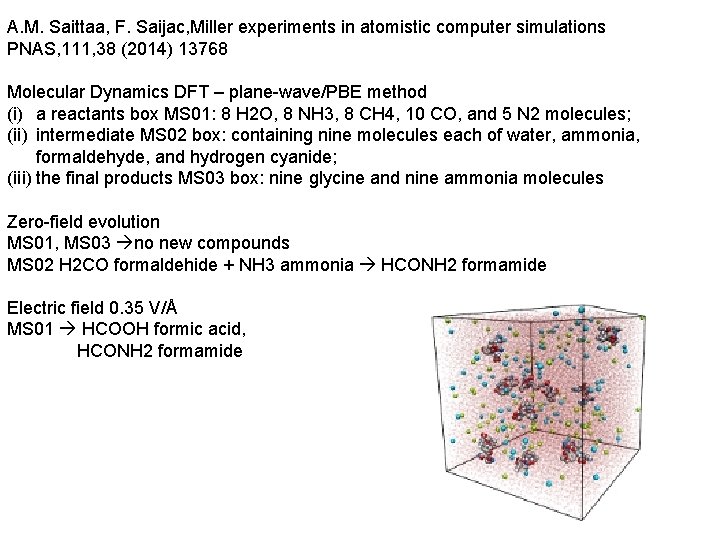
A. M. Saittaa, F. Saijac, Miller experiments in atomistic computer simulations PNAS, 111, 38 (2014) 13768 Molecular Dynamics DFT – plane-wave/PBE method (i) a reactants box MS 01: 8 H 2 O, 8 NH 3, 8 CH 4, 10 CO, and 5 N 2 molecules; (ii) intermediate MS 02 box: containing nine molecules each of water, ammonia, formaldehyde, and hydrogen cyanide; (iii) the final products MS 03 box: nine glycine and nine ammonia molecules Zero-field evolution MS 01, MS 03 no new compounds MS 02 H 2 CO formaldehide + NH 3 ammonia HCONH 2 formamide Electric field 0. 35 V/Å MS 01 HCOOH formic acid, HCONH 2 formamide

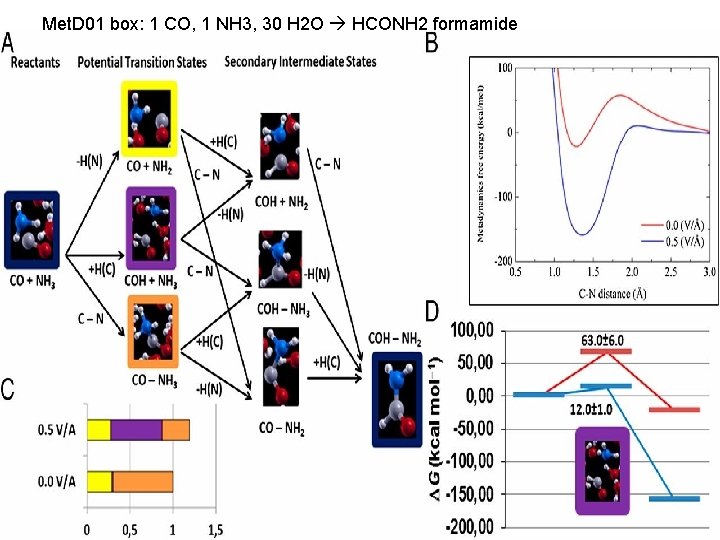
Met. D 01 box: 1 CO, 1 NH 3, 30 H 2 O HCONH 2 formamide

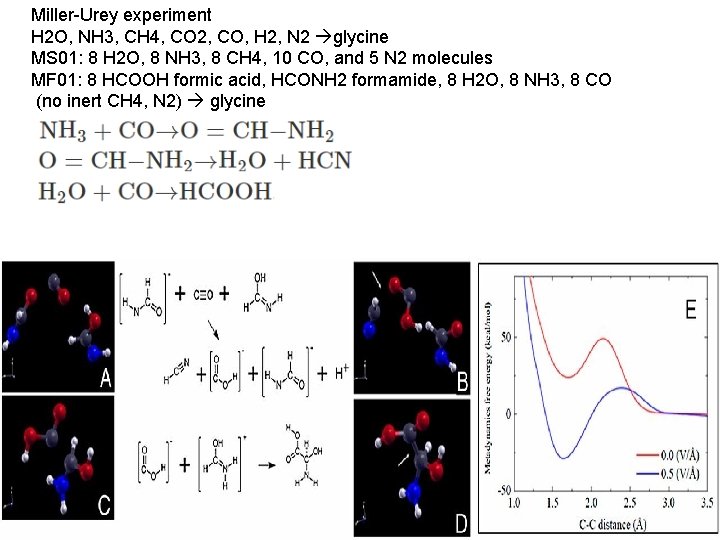
Miller-Urey experiment H 2 O, NH 3, CH 4, CO 2, CO, H 2, N 2 glycine MS 01: 8 H 2 O, 8 NH 3, 8 CH 4, 10 CO, and 5 N 2 molecules MF 01: 8 HCOOH formic acid, HCONH 2 formamide, 8 H 2 O, 8 NH 3, 8 CO (no inert CH 4, N 2) glycine

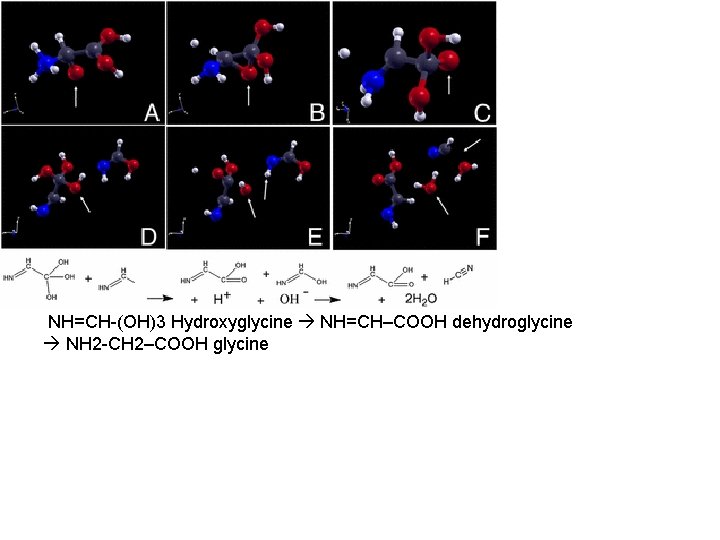
NH=CH-(OH)3 Hydroxyglycine NH=CH–COOH dehydroglycine NH 2 -CH 2–COOH glycine

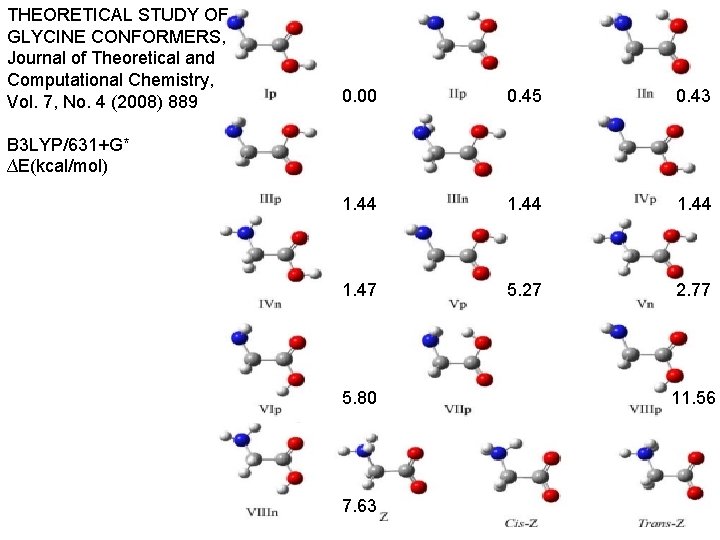
THEORETICAL STUDY OF GLYCINE CONFORMERS, Journal of Theoretical and Computational Chemistry, Vol. 7, No. 4 (2008) 889 0. 00 0. 45 0. 43 B 3 LYP/631+G* ∆E(kcal/mol) 1. 44 1. 44 1. 47 5. 27 2. 77 5. 80 11. 56 7. 63

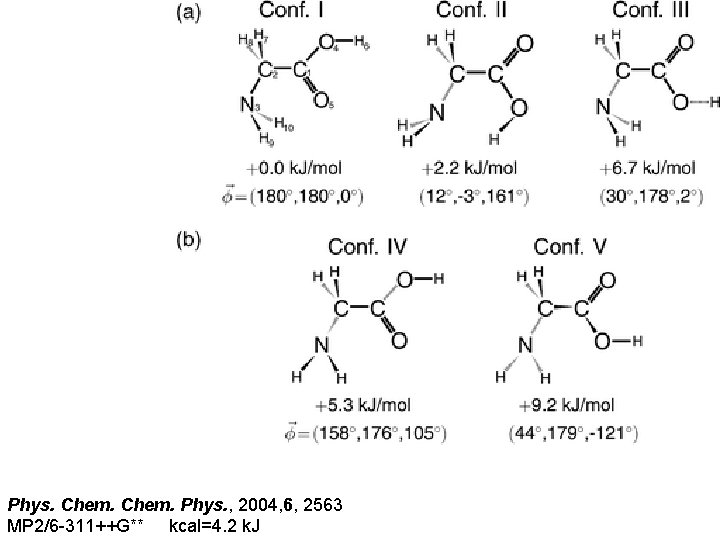
Phys. Chem. Phys. , 2004, 6, 2563 MP 2/6 -311++G** kcal=4. 2 k. J

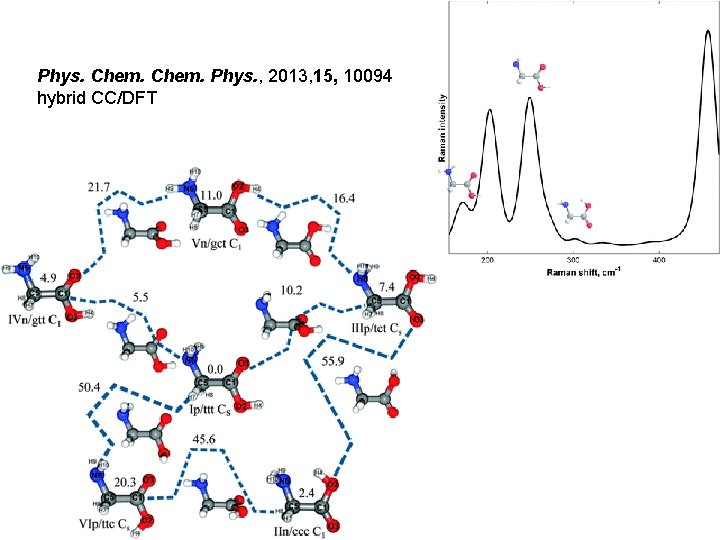
Phys. Chem. Phys. , 2013, 15, 10094 hybrid CC/DFT


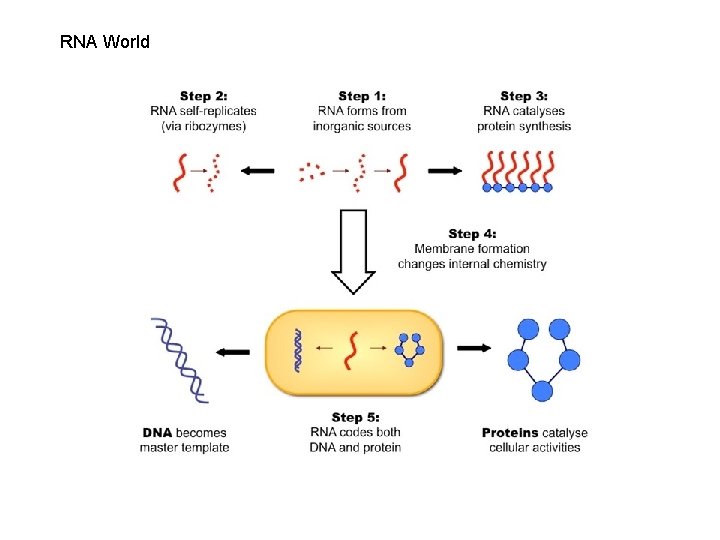
RNA World

Guanine, Adenine, and Hypoxanthine Production in UV-Irradiated Formamide Solutions: Relaxation of the Requirements for Prebiotic Purine Nucleobase Formation Chem. Bio. Chem 2010, 11, 1240

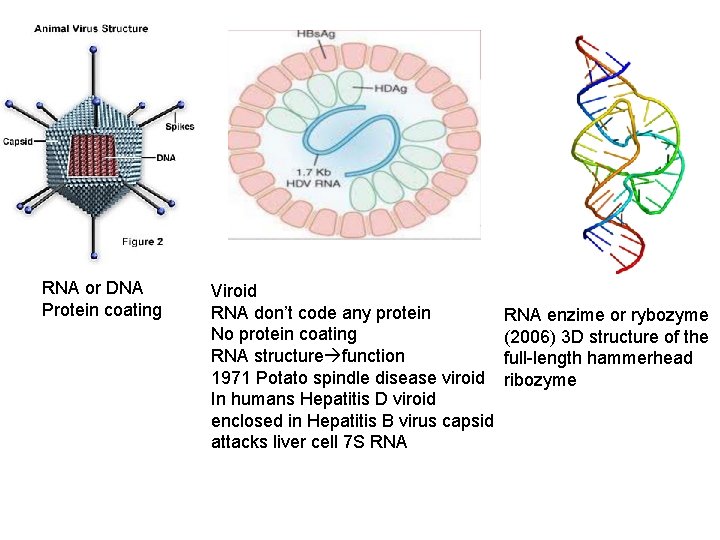
RNA or DNA Protein coating Viroid RNA don’t code any protein No protein coating RNA structure function 1971 Potato spindle disease viroid In humans Hepatitis D viroid enclosed in Hepatitis B virus capsid attacks liver cell 7 S RNA enzime or rybozyme (2006) 3 D structure of the full-length hammerhead ribozyme

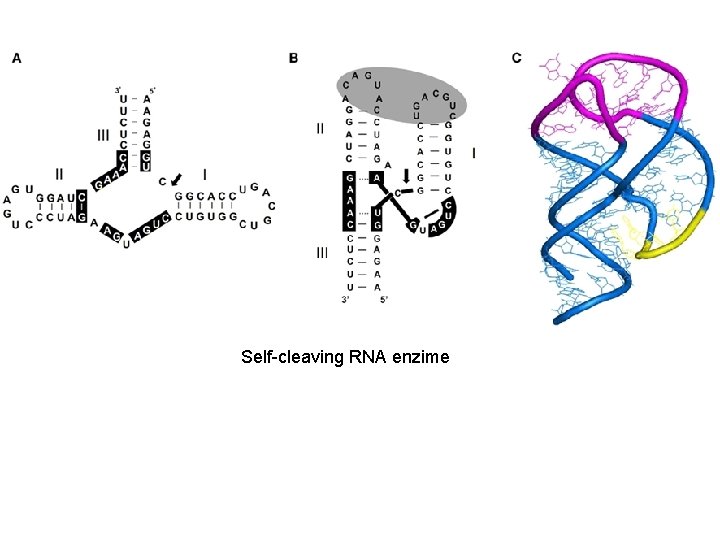
Self-cleaving RNA enzime

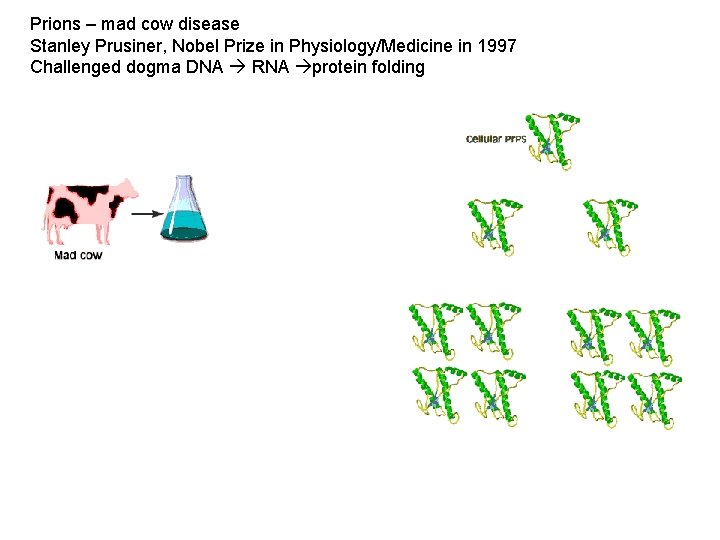
Prions – mad cow disease Stanley Prusiner, Nobel Prize in Physiology/Medicine in 1997 Challenged dogma DNA RNA protein folding

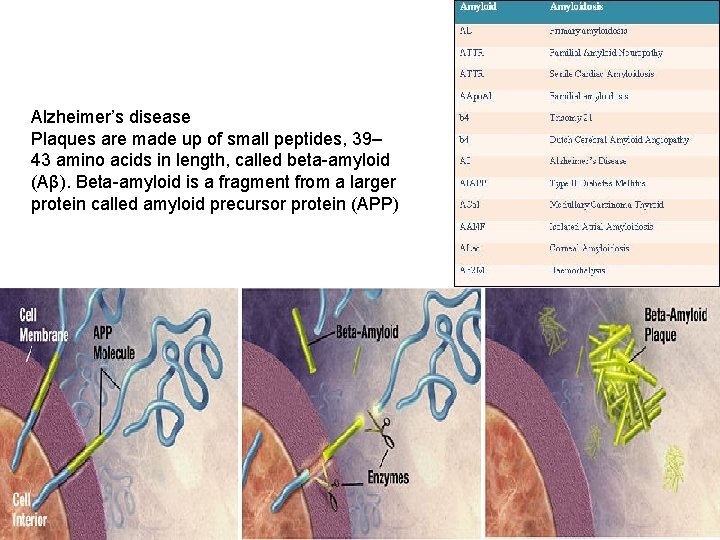
Alzheimer’s disease Plaques are made up of small peptides, 39– 43 amino acids in length, called beta-amyloid (Aβ). Beta-amyloid is a fragment from a larger protein called amyloid precursor protein (APP)




Amyloid β protein loop. The broken red lines indicate a loop in the amyloid-B protein that enables it to attach to other proteins and form clumps that kill brain cells.

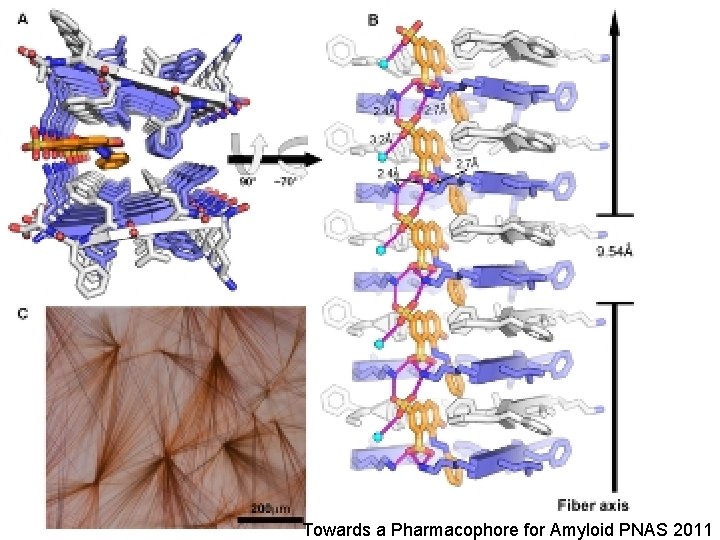
Towards a Pharmacophore for Amyloid PNAS 2011

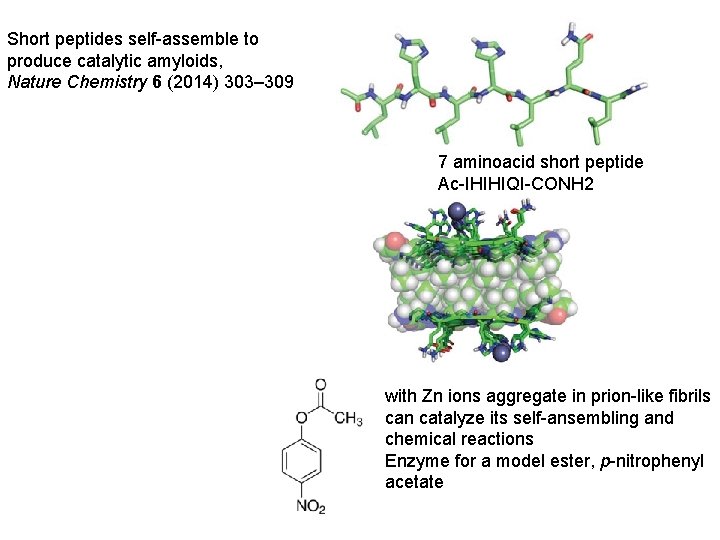
Short peptides self-assemble to produce catalytic amyloids, Nature Chemistry 6 (2014) 303– 309 7 aminoacid short peptide Ac-IHIHIQI-CONH 2 with Zn ions aggregate in prion-like fibrils can catalyze its self-ansembling and chemical reactions Enzyme for a model ester, p-nitrophenyl acetate

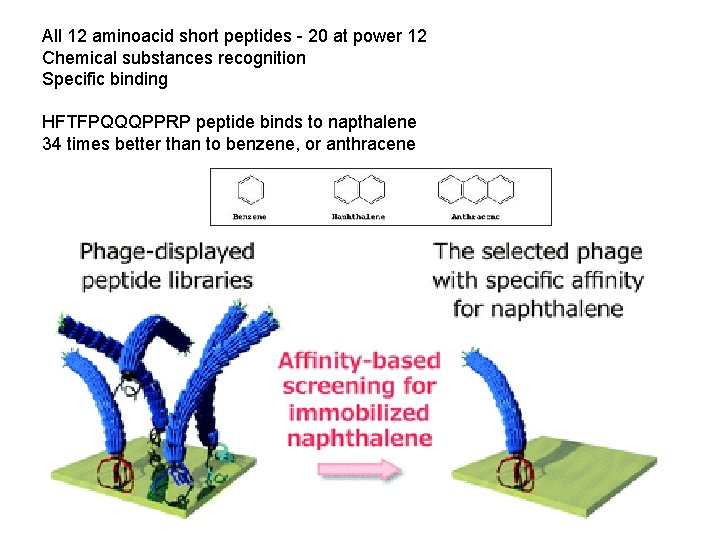
All 12 aminoacid short peptides - 20 at power 12 Chemical substances recognition Specific binding HFTFPQQQPPRP peptide binds to napthalene 34 times better than to benzene, or anthracene

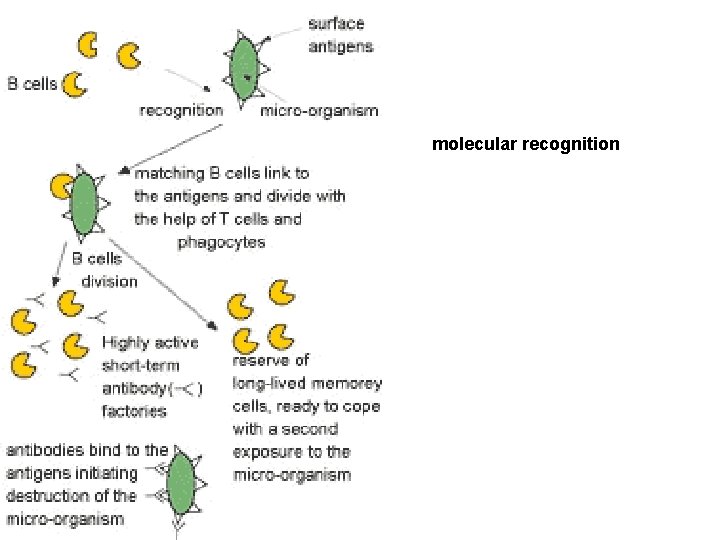
molecular recognition

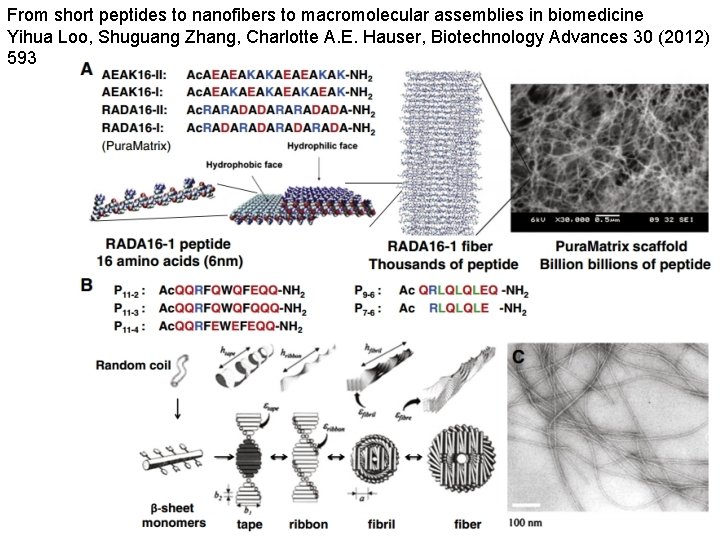
From short peptides to nanofibers to macromolecular assemblies in biomedicine Yihua Loo, Shuguang Zhang, Charlotte A. E. Hauser, Biotechnology Advances 30 (2012) 593


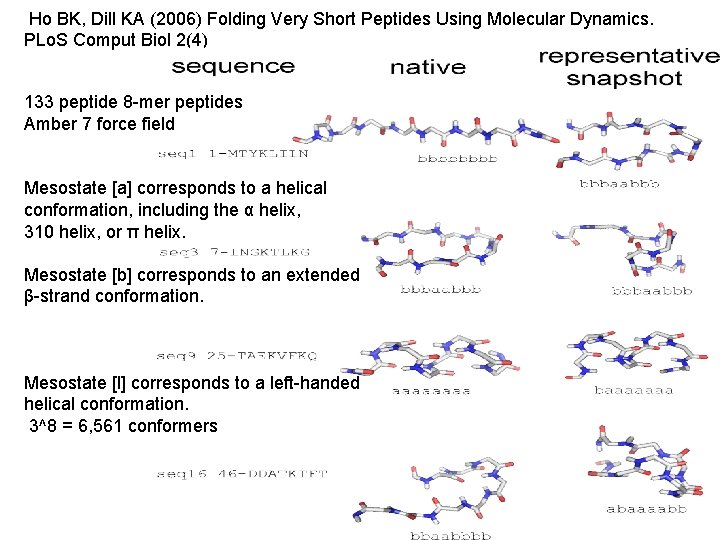
Ho BK, Dill KA (2006) Folding Very Short Peptides Using Molecular Dynamics. PLo. S Comput Biol 2(4) 133 peptide 8 -mer peptides Amber 7 force field Mesostate [a] corresponds to a helical conformation, including the α helix, 310 helix, or π helix. Mesostate [b] corresponds to an extended β-strand conformation. Mesostate [l] corresponds to a left-handed helical conformation. 3^8 = 6, 561 conformers

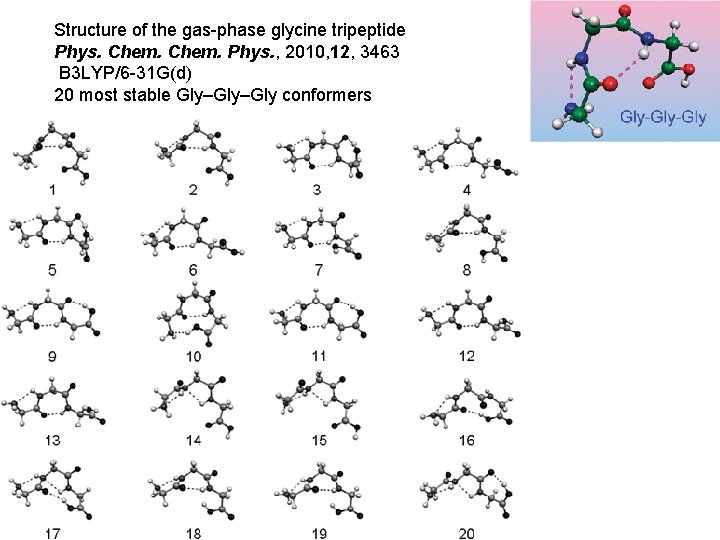
Structure of the gas-phase glycine tripeptide Phys. Chem. Phys. , 2010, 12, 3463 B 3 LYP/6 -31 G(d) 20 most stable Gly–Gly conformers
 The crucible 1953
The crucible 1953 1924-1953
1924-1953 Cherry 1953 dichotic listening
Cherry 1953 dichotic listening Alignment with west 1953-62
Alignment with west 1953-62 Napoleon abueva kaganapan sculpture
Napoleon abueva kaganapan sculpture Modal model of memory
Modal model of memory La maxima tension 1947 y 1953
La maxima tension 1947 y 1953 Negative priming
Negative priming Donald broadbent theory
Donald broadbent theory Arthur millers drama fra 1953
Arthur millers drama fra 1953 Recommendation of secondary education commission 1952-53
Recommendation of secondary education commission 1952-53 Fahrenheit 451 1953
Fahrenheit 451 1953 War of the worlds 1953
War of the worlds 1953 2007-1953
2007-1953 Fhrer
Fhrer 1953
1953 Red china johnnie ray
Red china johnnie ray July 26 1953
July 26 1953 The origins of sociology
The origins of sociology Selection and gradation in language teaching
Selection and gradation in language teaching The magic of kefir
The magic of kefir The hunger games questions
The hunger games questions Historical origins of the health belief model
Historical origins of the health belief model Rastafarian origin
Rastafarian origin Cwv-101 origins
Cwv-101 origins Origins of the cold war
Origins of the cold war Origins of the cold war
Origins of the cold war History of weaving
History of weaving Origins of judo
Origins of judo Origins of tennis
Origins of tennis Hamstring insertions and origins
Hamstring insertions and origins Words with the root am
Words with the root am Sikhism place of origin
Sikhism place of origin