Open Dose on EGI producing reference dosimetric data
















- Slides: 35

Open. Dose on EGI: producing reference dosimetric data for Nuclear Medicine on the European Grid Infrastructure Gilles Mathieu (1), Maxime Chauvin (2), Axel Bonnet (3), Sorina Camarasu Pop (3), Isabelle Perseil (1), Manuel Bardiès (2) (1) Inserm, DSI, Coordination de l’Informatique Scientifique, Paris, France (2) CRCT, UMR 1037, INSERM, Université Toulouse III Paul Sabatier, Toulouse, France (3) CREATIS, CNRS, INSERM, Université Claude Bernard Lyon I, INSA, Lyon, France Open. Gate meeting, December 13 th 2018, Orsay

PLAN A PLAN B • Scientific view • Technical view • Foreseen solution • Start the talk • Infrastructures • See how it goes • Tools • ☺ • First results • Conclusion Open. Dose – Open. Gate 2018 13/12/2018 2

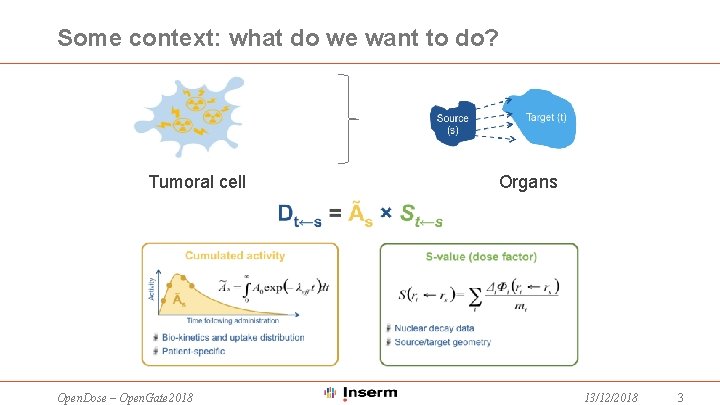
Some context: what do we want to do? Tumoral cell Open. Dose – Open. Gate 2018 Organs 13/12/2018 3

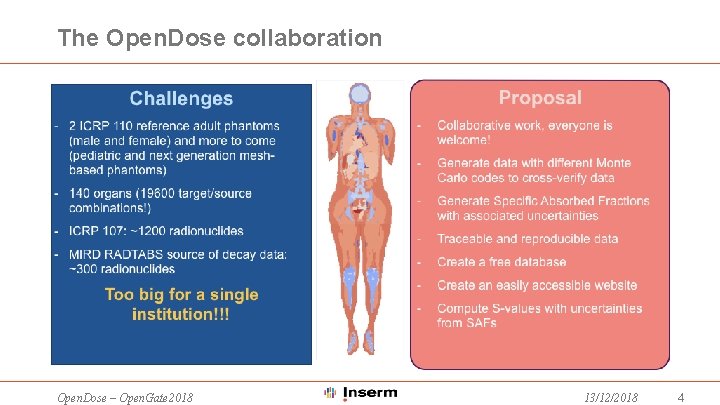
The Open. Dose collaboration Open. Dose – Open. Gate 2018 13/12/2018 4

And what’s the matter? • 1 simulation = 1 Gate/Geant 4 job • Gate -a [Source_ID, 95][particle, gamma][energy, 1][nb, 10 e 5] main_AF. mac • 50 960 simulations • 140 source organs, 2 particle types, 91 energy levels, 2 models • On a local cluster (240 CPU) : • 1 organ x 2 particles x 91 energies x 1 model = 1 day More than 1 million CPU hours Open. Dose – Open. Gate 2018 13/12/2018 5

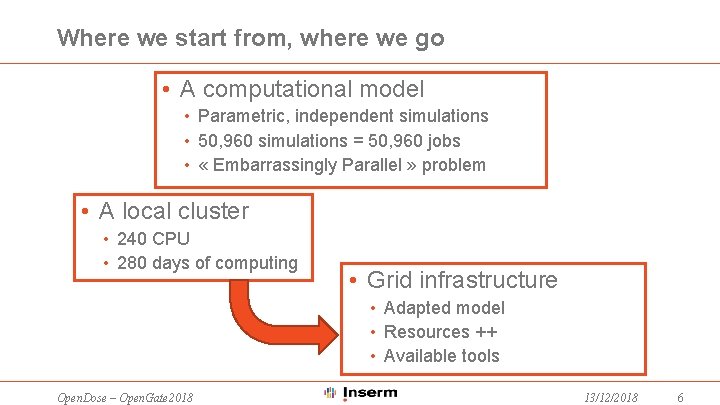
Where we start from, where we go • A computational model • Parametric, independent simulations • 50, 960 simulations = 50, 960 jobs • « Embarrassingly Parallel » problem • A local cluster • 240 CPU • 280 days of computing • Grid infrastructure • Adapted model • Resources ++ • Available tools Open. Dose – Open. Gate 2018 13/12/2018 6

Solution • Infrastructure Together, we’ll be many! • Tools Gate/Geant 4 on the grid Open. Dose – Open. Gate 2018 13/12/2018 7

Open. Dose – Open. Gate 2018 13/12/2018 8

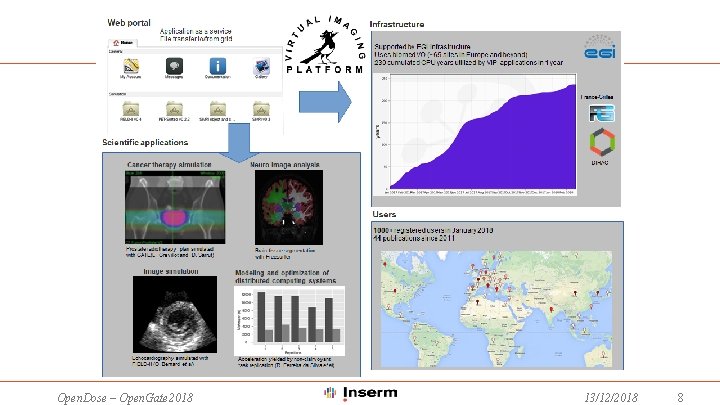
Gate. Lab allows you to submit your GATE simulations on EGI from a simple web page: • • Upload your data and main macro file from the web browser. Select the estimated CPU time needed to complete your simulation. Choose the GATE release. After completion, results are automatically merged and ready for download. Open. Dose requires the new Dose. By. Regions output: Need to update the GATE release to the latest version 8. 1 Need to update the merger Open. Dose – Open. Gate 2018 13/12/2018 9

Gate. Lab • Update of the GATE release • • • Docker. File which build a new image with Geant 4, ROOT and GATE on Cent. OS. Script to extract the GATE binary and dependencies from the Docker image. Add a env. sh to ease the setup of Geant 4 and ROOT environment variables. GATE release 8. 1 is now available • Update of the merger for the Dose. By. Regions output • Almost finished. Open. Dose – Open. Gate 2018 13/12/2018 10

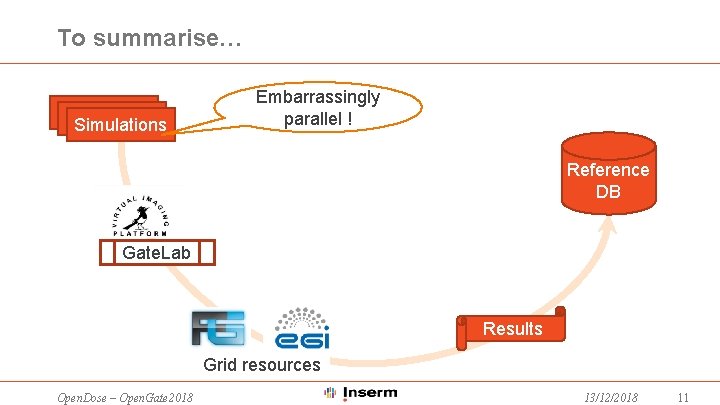
To summarise… Simulations Embarrassingly parallel ! Reference DB Gate. Lab Results Grid resources Open. Dose – Open. Gate 2018 13/12/2018 11

Nice. However… • Generate 50 960 configuration files? NEIN ! Thou shalt use parameters A NEW WORKFLOW • Manually launch 50, 960 jobs through the portal? NEIN Open. Dose – Open. Gate 2018 ! Thou shalt script A CLIENT CODE 13/12/2018 12

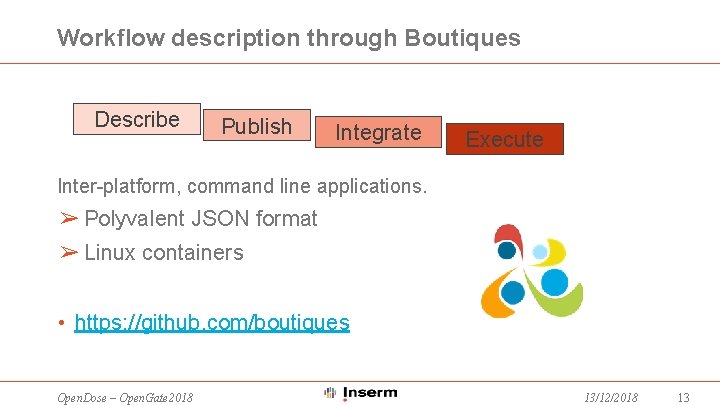
Workflow description through Boutiques Describe Publish Integrate Execute Inter-platform, command line applications. ➢ Polyvalent JSON format ➢ Linux containers • https: //github. com/boutiques Open. Dose – Open. Gate 2018 13/12/2018 13

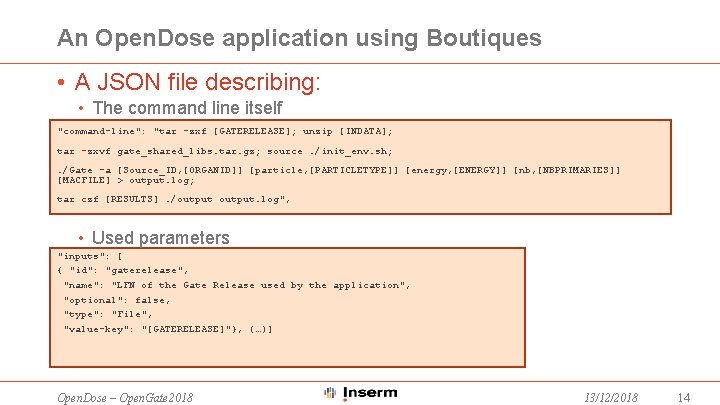
An Open. Dose application using Boutiques • A JSON file describing: • The command line itself "command-line": "tar -zxf [GATERELEASE]; unzip [INDATA]; tar -zxvf gate_shared_libs. tar. gz; source. /init_env. sh; . /Gate -a [Source_ID, [ORGANID]] [particle, [PARTICLETYPE]] [energy, [ENERGY]] [nb, [NBPRIMARIES]] [MACFILE] > output. log; tar czf [RESULTS]. /output. log", • Used parameters "inputs": [ { "id": "gaterelease", "name": "LFN of the Gate Release used by the application", "optional": false, "type": "File", "value-key": "[GATERELEASE]"}, (…)] Open. Dose – Open. Gate 2018 13/12/2018 14

An Open. Dose application using Boutiques Open. Dose – Open. Gate 2018 13/12/2018 15

The CARMIN API • https: //github. com/CARMIN-org • https: //app. swaggerhub. com/apis/CARMIN Open. Dose – Open. Gate 2018 13/12/2018 16

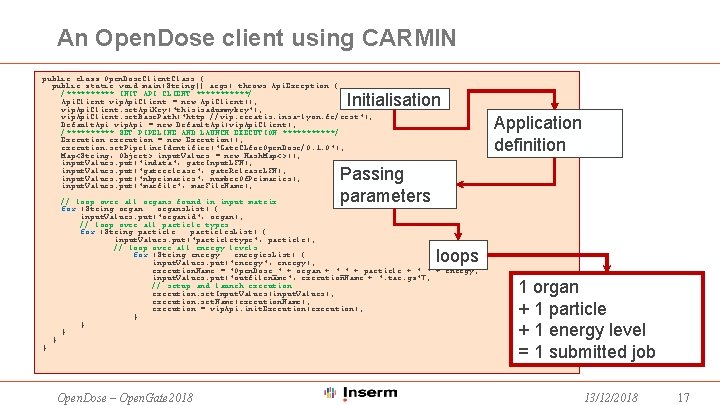
An Open. Dose client using CARMIN public class Open. Dose. Client. Class { public static void main(String[] args) throws Api. Exception { /***** INIT API CLIENT ******/ Api. Client vip. Api. Client = new Api. Client(); vip. Api. Client. set. Api. Key("thisisadummykey"); vip. Api. Client. set. Base. Path("http: //vip. creatis. insa-lyon. fr/rest"); Default. Api vip. Api = new Default. Api(vip. Api. Client); /***** SET PIPELINE AND LAUNCH EXECUTION ******/ Execution execution = new Execution(); execution. set. Pipeline. Identifier("Gate. CLfor. Open. Dose/0. 1. 0"); Map<String, Object> input. Values = new Hash. Map<>(); input. Values. put("indata", gate. Input. LFN); input. Values. put("gaterelease", gate. Release. LFN); input. Values. put("nbprimaries", number. Of. Primaries); input. Values. put("macfile", mac. File. Name); Initialisation Application definition Passing parameters // loop over all organs found in input matrix for (String organ : organs. List) { input. Values. put("organid", organ); // loop over all particle types for (String particle : particles. List) { input. Values. put("particletype", particle); // loop over all energy levels for (String energy : energies. List) { input. Values. put("energy", energy); execution. Name = "Open. Dose_" + organ + "_" + particle + "_" + energy; input. Values. put("outfilename", execution. Name + ". tar. gz"); // setup and launch execution. set. Input. Values(input. Values); execution. set. Name(execution. Name); execution = vip. Api. init. Execution(execution); } } } loops } } Open. Dose – Open. Gate 2018 1 organ + 1 particle + 1 energy level = 1 submitted job 13/12/2018 17

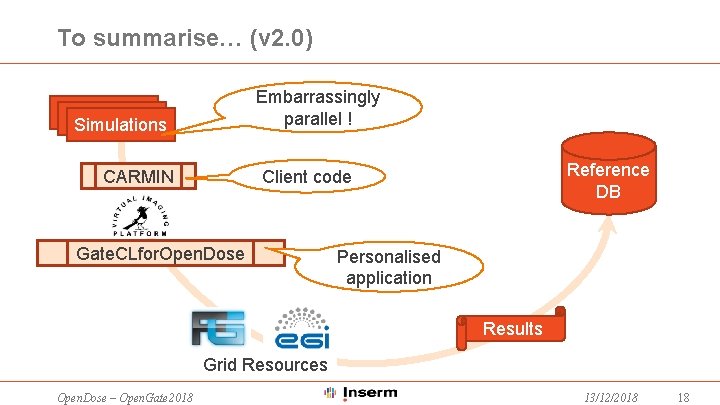
To summarise… (v 2. 0) Embarrassingly parallel ! Simulations Reference DB Client code CARMIN Gate. CLfor. Open. Dose Personalised application Results Grid Resources Open. Dose – Open. Gate 2018 13/12/2018 18

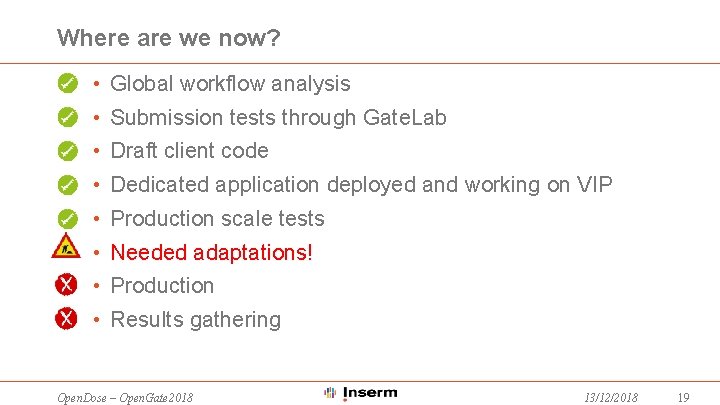
Where are we now? • Global workflow analysis • Submission tests through Gate. Lab • Draft client code • Dedicated application deployed and working on VIP • Production scale tests • Needed adaptations! • Production • Results gathering Open. Dose – Open. Gate 2018 13/12/2018 19

Needed adaptations ? • Long jobs (>24 h) don’t run well on the grid • GOOD NEWS: Gate. Lab has a mechanism to split jobs • BAD NEWS: Gate. CLfor. Open. Dose doesn’t Open. Dose – Open. Gate 2018 13/12/2018 20

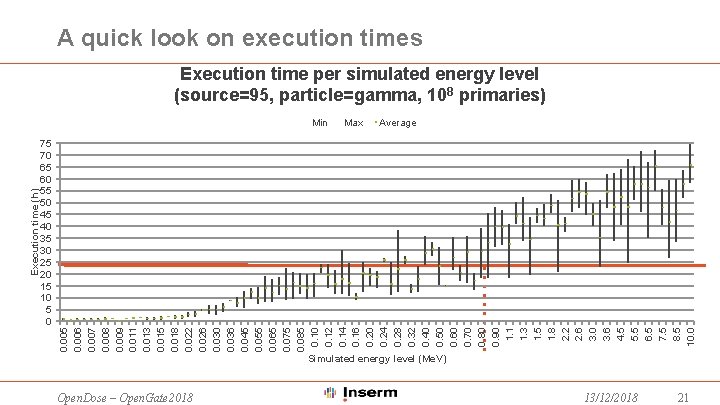
A quick look on execution times Execution time per simulated energy level (source=95, particle=gamma, 108 primaries) 75 70 65 60 55 50 45 40 35 30 25 20 15 10 5 0 Max Average 0. 005 0. 006 0. 007 0. 008 0. 009 0. 011 0. 013 0. 015 0. 018 0. 022 0. 026 0. 030 0. 036 0. 045 0. 055 0. 065 0. 075 0. 085 0. 10 0. 12 0. 14 0. 16 0. 20 0. 24 0. 28 0. 32 0. 40 0. 50 0. 60 0. 70 0. 80 0. 90 1. 1 1. 3 1. 5 1. 8 2. 2 2. 6 3. 0 3. 6 4. 5 5. 5 6. 5 7. 5 8. 5 10. 0 Execution time (h) Min Simulated energy level (Me. V) Open. Dose – Open. Gate 2018 13/12/2018 21

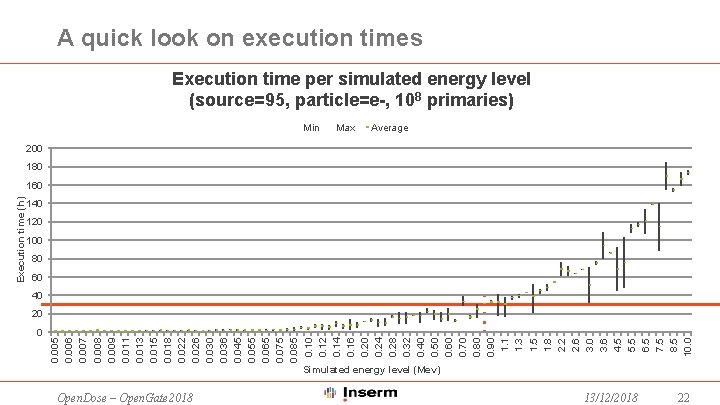
A quick look on execution times Execution time per simulated energy level (source=95, particle=e-, 108 primaries) Min Max Average 200 180 140 120 100 80 60 40 20 0 0. 005 0. 006 0. 007 0. 008 0. 009 0. 011 0. 013 0. 015 0. 018 0. 022 0. 026 0. 030 0. 036 0. 045 0. 055 0. 065 0. 075 0. 085 0. 10 0. 12 0. 14 0. 16 0. 20 0. 24 0. 28 0. 32 0. 40 0. 50 0. 60 0. 70 0. 80 0. 90 1. 1 1. 3 1. 5 1. 8 2. 2 2. 6 3. 0 3. 6 4. 5 5. 5 6. 5 7. 5 8. 5 10. 0 Execution time (h) 160 Simulated energy level (Mev) Open. Dose – Open. Gate 2018 13/12/2018 22

Houston, we have a problem • Only 2/3 of the overseen production can run smoothly in current situation • Simulations with energy level < 0. 8 Mev • We need to either: • Implement a split mechanism in our application • Modify Gate. Lab • WORK IN PROGRESS! Keep tuned ; -) Open. Dose – Open. Gate 2018 13/12/2018 23

Meanwhile… • We have started with REAL production anyway • All source organs, e- and gamma • Male and Female model • Submission started Tuesday afternoon (11. Dec. ) • Around 2000 simulations finished, and counting

A first feedback The light side The not-so-light side • Existing tools • Efficacient co-workers • Need for fine tuning • Slow start • Automation • Optimisation • Specific development means we may lose benefits of existing tools Open. Dose – Open. Gate 2018 13/12/2018 25

To the future, and beyond • Industrialisation • Optimise code and submission • Use more/different models • Ensure reproducibility et traceability • Scientific exploration • Re-use submission framework • Test other MC softwares • Technical Exploration • Other tools? • Other infrastructures? Open. Dose – Open. Gate 2018 13/12/2018 26

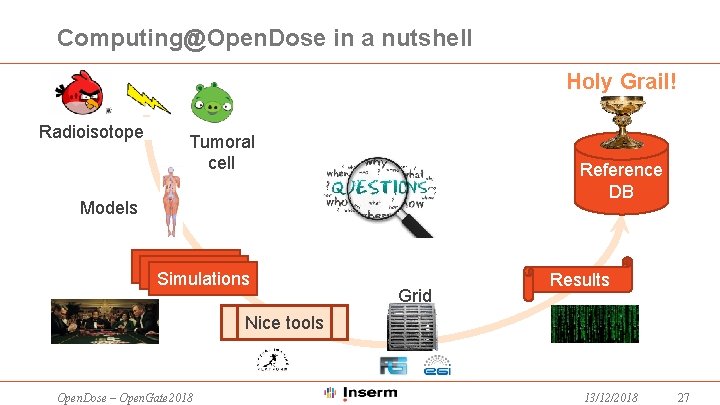
Computing@Open. Dose in a nutshell Holy Grail! Radioisotope Tumoral cell Reference DB Models Simulations Grid Results Nice tools Open. Dose – Open. Gate 2018 13/12/2018 27

Annexes Informations, links & references for an offline reading

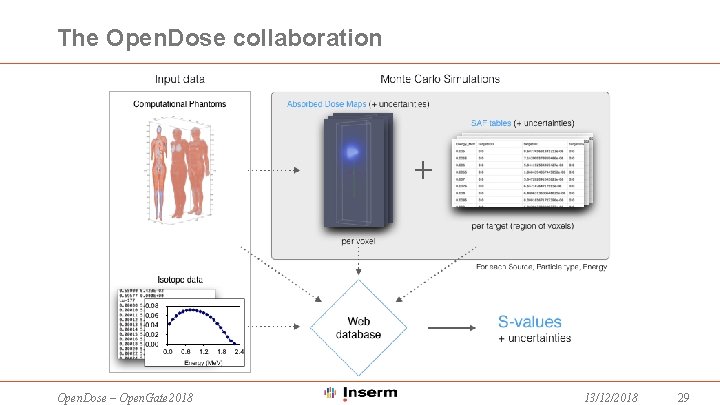
The Open. Dose collaboration Open. Dose – Open. Gate 2018 13/12/2018 29

More on Open. Dose • MCMA 2017 talk by M. Chauvin • http: //people. na. infn. it/~mettivie/MCMA%20 presentation/18%20 Aul a%20 Magna/9_45_Chauvin. pdf • MRTDosimetry 2018 talk by M. Chauvin • http: //mrtdosimetry-empir. eu/wpcontent/uploads/2018/10/13_MRTdosimetry. Workshop_Chauvin. pd f Open. Dose – Open. Gate 2018 13/12/2018 30

France Grilles • Une offre de services sur une infrastructure informatique distribuée pour le calcul et le stockage de données scientifiques • Une équipe d’animation au service de l’ensemble des acteurs qui coordonne les opérations de cette infrastructure basée sur les technologies de grille et de cloud informatiques • Une communauté de plus d’un millier d’utilisateurs dynamiques, prêts à partager leurs connaissances et à s’entraider • Le représentant de la France au niveau international au sein d’EGI • Et surtout et avant tout un réseau résolument humain ! • www. france-grilles. fr Open. Dose – Open. Gate 2018 13/12/2018 31

EGI • www. egi. eu Open. Dose – Open. Gate 2018 13/12/2018 32

Boutiques • Décrire, publier, intégrer et exécuter des applications en ligne de commande inter-plateformes. • faciliter le portage d'applications • import et échange d'applications • science ouverte et reproductible • Format JSON polyvalent pour décrire la ligne de commande, les entrées et les sorties • Utilisation de conteneurs Linux pour faciliter l'installation et le partage d'applications • https: //github. com/boutiques Open. Dose – Open. Gate 2018 13/12/2018 33

Portage des jobs Open. Dose sur la grille • Code client pour utiliser VIP via CARMIN • https: //github. com/CISI-INSERM/Open. Dose. Computing/tree/master/VIPclient • Descripteur JSON pour portage de la ligne de commande Open. Dose dans VIP via Boutiques • https: //github. com/CISI-INSERM/Open. Dose. Computing/tree/master/Boutiques Open. Dose – Open. Gate 2018 13/12/2018 34

Crédits • CRCT, UMR 1037, INSERM, Université Toulouse III Paul Sabatier, Toulouse, France • Maxime Chauvin, Manuel Bardiès • CREATIS, CNRS, INSERM, Université Claude Bernard Lyon I, INSA, Lyon, France • Axel Bonnet, Sorina Camarasu Pop • INSERM, DSI, Coordination de l’Informatique Scientifique, Paris, France • Gilles Mathieu, Isabelle Perseil Open. Dose – Open. Gate 2018 13/12/2018 35