Notes on HYSPLIT 4SV Simulations Mexico City Workshop





















- Slides: 52

Notes on HYSPLIT 4_SV Simulations Mexico City Workshop, Feb 2012 Mark Cohen NOAA Air Resources Laboratory, Silver Spring, MD, USA Paul Bartlett St. Peters College, Newark, NJ, USA

HYSPLIT-format meteorological data prepared for these simulations 2/10/2012 Mexico City Workshop, Feb 2012

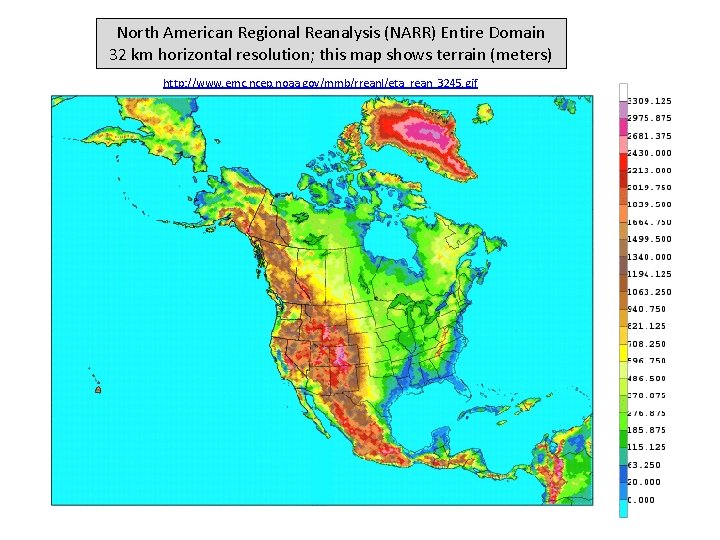
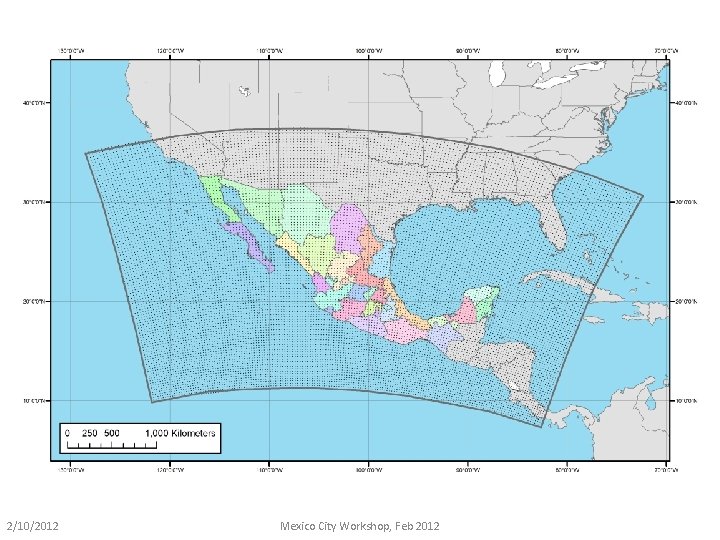
North American Regional Reanalysis (NARR) Entire Domain 32 km horizontal resolution; this map shows terrain (meters) http: //www. emc. ncep. noaa. gov/mmb/rreanl/eta_rean_3245. gif

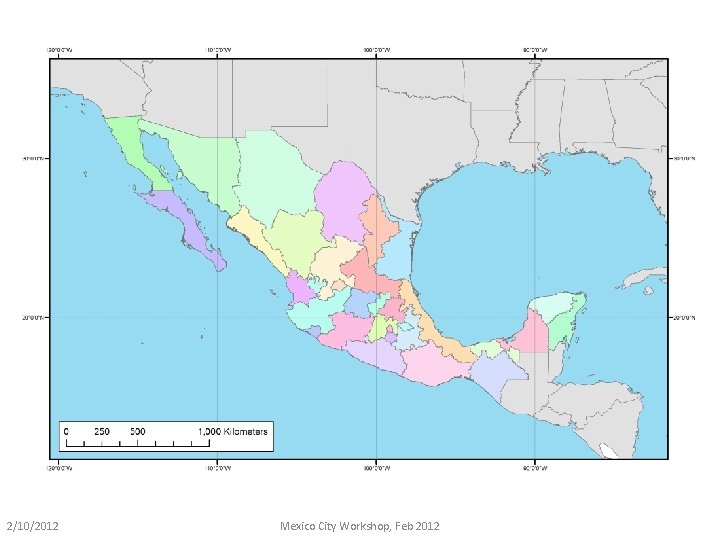
36 km North American Regional Reanalysis (NARR) • Subgrid (shown below) extracted from full NARR grid for this project • Why a subgrid? Less data to transfer & computational efficiency • July 2007 – Jan 2011 (for simulations from 2008 -2010) • Converted to HYSPLIT format • Monthly files with 3 hr temporal resolution • Each monthly file ~650 MB • 43 months (July 2007 – Jan 2011) ~ 27 GB 2/10/2012 Mexico City Workshop, Feb 2012

2/10/2012 Mexico City Workshop, Feb 2012

2/10/2012 Mexico City Workshop, Feb 2012

2/10/2012 Mexico City Workshop, Feb 2012

2/10/2012 Mexico City Workshop, Feb 2012

TO DO TEST RUNS, MUST INSTALL “basic” HYSPLIT first, so that you have graphical utilities, etc. 2/10/2012 Mexico City Workshop, Feb 2012

An installation package for “basic” HYSPLIT has been placed on the FTP site under the following folder: PLAIN_HYSPLIT 4_INSTALLATION To do the various installations, you will need administrative privileges on the computer. Use default install locations if you can. First install utilities, in the following order. TCL (Active. Tcl 8. 5. 8. 1. 291945 -win 32 -ix 86 -threaded. exe) Ghostscript (gs 863 w 32. exe) Ghostview (gsv 49 w 32. exe) Image. Magick (Image. Magick-6. 6. 1 -5 -Q 8 -windows-dll. exe) Then install the HYSPLIT program (HYSPLIT_win 32 R. exe) You can always download latest PC version from: https: //ready. arl. noaa. gov/HYSPLIT. php 2/10/2012 Mexico City Workshop, Feb 2012

First, try to do one initial test Run 2/10/2012 Mexico City Workshop, Feb 2012

1. USING BINARY MODE, download folder from the FTP site: hysplit 4_SV_feb_2012 2. Open up a DOS Window 3. Navigate to c: hysplit 4_SV_feb_2012working_SV 4. To see batch files present, type: dir *. bat [return] 5. Install Notepad++ or Some Other Text Editor with Context-Highlighting 6. Examine SET_SV_007. bat [open it up in an editor; don’t double click on it!] 7. Adjust the “environment” [ENVR==2] … see the next page 8. Once you’ve done this, return to the DOS window (working_SV) 9. Try to run the first test by typing: run_test_1 then hit return 10. If it all works, you will see the program running, and when done, you will end up with the following directory: C: hysplit 4_SV_feb_2012HYSPLIT_RESULTStestingtest_run_1 11. Navigate to this directory from Windows Explorer and examine the files there 2/10/2012 Mexico City Workshop, Feb 2012

For your installation, the directories may be the same as shown below. But if they are not, edit them the text below to correspond to your situation rem ************************************ if %ENVR%==2 ( rem right now same as ENVR=1, but, can be customized for any given local rem environment, on anyone's particular computer rem ************************************ rem directory for overall run and set batch files (do NOT include back slash at end) SET master_work_dir=C: hysplit 4_SV_feb_2012working_SV rem here is the base directory for met data; individual datasets are subdirectories of this rem met data home directory - do NOT include backslash, will be included later SET met_home=C: hysplit 4_SV_feb_2012metdata rem reaction concentration data directory (DO include back slash at end) SET conc_data=C: hysplit 4_SV_feb_2012gridded_chem_data1999_global_EMEP_data rem directory for concplot, con 2 asc (do NOT include back slash at end) SET conc_exec_dir=C: hysplit 4_SV_feb_2012exec_release_v 23 a if %debug_release%==debug SET hycs_exec_dir=C: hysplit 4_SV_feb_2012exec_debug_v 23 a rem HYPSLIT-SV executable directory (do NOT include back slash at end) if %debug_release%==release SET hycs_exec_dir=C: hysplit 4_SV_feb_2012exec_release_v 23 a rem HYPSLIT-SV executable directory (do NOT include back slash at end) ) rem ************* rem end of ENVR=2 rem ************* 2/10/2012 Mexico City Workshop, Feb 2012

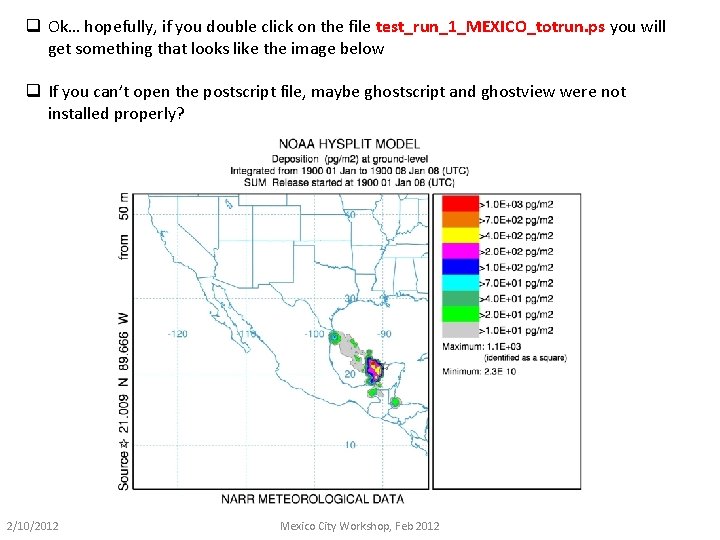
q Ok… hopefully, if you double click on the file test_run_1_MEXICO_totrun. ps you will get something that looks like the image below q If you can’t open the postscript file, maybe ghostscript and ghostview were not installed properly? 2/10/2012 Mexico City Workshop, Feb 2012

There a lot of other files, and we will examine them shortly… But for now, lets just make sure that this initial run worked. If you got the postscript image on the previous page, it worked! If not, we will have to figure out what is going on… Don’t worry, usually the problems are relatively easy to find and fix. A “pre-computed” version of the output from this test is provided, in the directory “pre_computed_testing” 2/10/2012 Mexico City Workshop, Feb 2012

HYSPLIT produces output as postscript files (. ps). These can be viewed with Ghost. View, but they are not very convenient to use. For example, they can’t easily be imported into Power. Point or MS-Word. If you have Image. Magick installed, you can “convert” the postscript file to a jpg (or other format) using the following command from the DOS command line: convert -density 300 test_run_1_MEXICO_totrun. ps name. jpg (where “name” can be anything you want… ) If the program “convert” was not found, check your PATH, and either change your PATH or copy the program “convert. exe” from the Image. Magick directory to someplace in your PATH. On my computer, the “convert” program is at the following location: C: Program Files (x 86)Image. Magick-6. 6. 1 -Q 8convert. exe 2/10/2012 Mexico City Workshop, Feb 2012

IMPORTANT NOTE ABOUT MET DATA In order to minimize download time and facilitate initial testing, only a minimum set of met files have been included in the hysplit 4_SV_feb_2012 folder that you have downloaded from the FTP site. In fact, only “two” files have been included: One NARR_MX_36 km file, for Jan 2008: C: hysplit 4_SV_feb_2012metdataNARR_MX_36 km NARR_MX_2008_JAN. bin And one NCEP/NCAR Global Reanalysis file, for Jan 2008: C: hysplit 4_SV_feb_2012metdataGLOBAL_2 p 5 RP 200801. gbl Additional files are available on the FTP site, and in order to carry out runs for time periods other than Jan 2008, you can download these from the FTP site. Remember to use BINARY MODE. You should put any additional files you download in the same folders as the two files above. The full set of NARR_MX_36 km met files is in the FTP folder: NARR_MX_met_data The full set of Global_Reanalysis met files is in the FTP folder: Global_MX_met_data 2/10/2012 Mexico City Workshop, Feb 2012

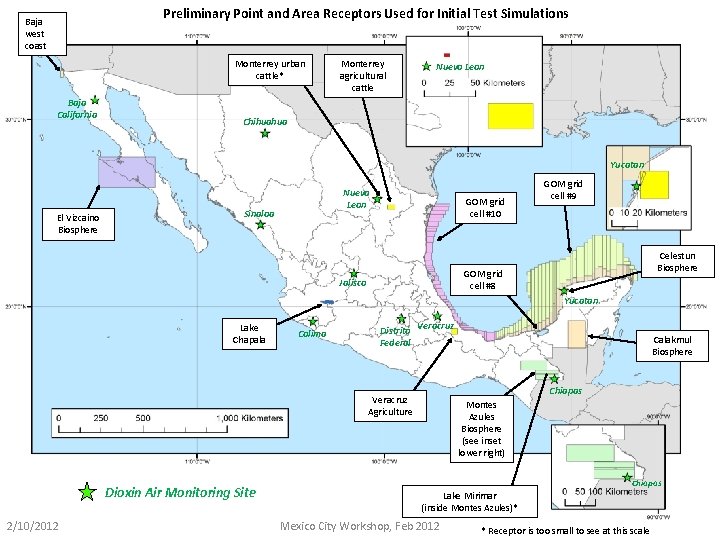
Receptors Used for Initial Test Simulations (on the following page is a map of the receptors that we have set up so far) 2/10/2012 Mexico City Workshop, Feb 2012

Preliminary Point and Area Receptors Used for Initial Test Simulations Baja west coast Monterrey urban cattle* Baja California Monterrey agricultural cattle Nuevo Leon Chihuahua Yucatan El Vizcaino Biosphere Nuevo Leon Sinaloa GOM grid cell #10 GOM grid cell #9 Celestun Biosphere GOM grid cell #8 Jalisco Yucatan Lake Chapala Colima Distrito Federal Veracruz Calakmul Biosphere Chiapas Veracruz Agriculture Dioxin Air Monitoring Site 2/10/2012 Montes Azules Biosphere (see inset lower right) Chiapas Lake Mirimar (inside Montes Azules)* Mexico City Workshop, Feb 2012 * Receptor is too small to see at this scale

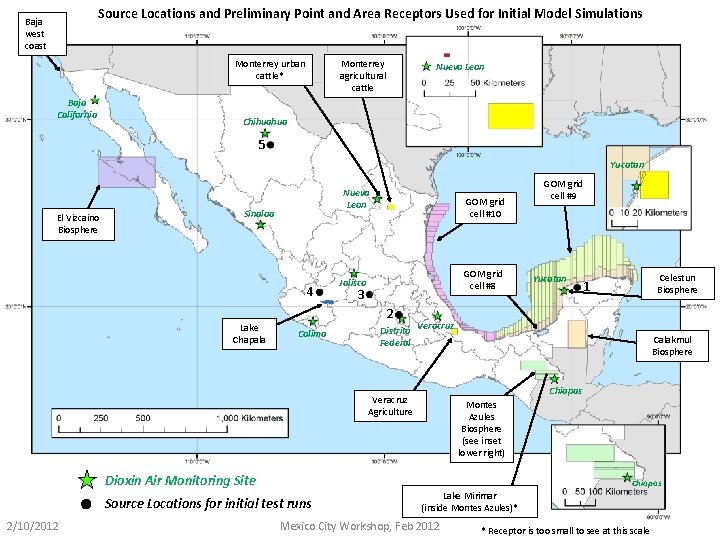
Source Locations and Preliminary Point and Area Receptors Used for Initial Model Simulations Baja west coast Monterrey agricultural cattle Monterrey urban cattle* Baja California Nuevo Leon Chihuahua 5 Yucatan El Vizcaino Biosphere Nuevo Leon Sinaloa 4 Lake Chapala GOM grid cell #10 GOM grid cell #8 Jalisco 3 2 Colima Distrito Federal Yucatan Celestun Biosphere 1 Veracruz Calakmul Biosphere Chiapas Veracruz Agriculture Montes Azules Biosphere (see inset lower right) Dioxin Air Monitoring Site Chiapas Source Locations for initial test runs 2/10/2012 GOM grid cell #9 Lake Mirimar (inside Montes Azules)* Mexico City Workshop, Feb 2012 * Receptor is too small to see at this scale

SUMMARY OF FILES PRODUCED DURING EACH SIMULATION 2/10/2012 Mexico City Workshop, Feb 2012

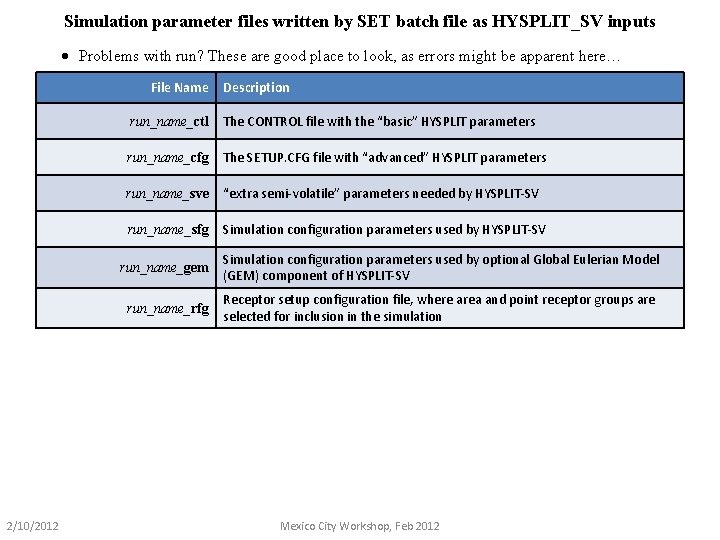
Simulation parameter files written by SET batch file as HYSPLIT_SV inputs · Problems with run? These are good place to look, as errors might be apparent here… File Name 2/10/2012 Description run_name_ctl The CONTROL file with the “basic” HYSPLIT parameters run_name_cfg The SETUP. CFG file with “advanced” HYSPLIT parameters run_name_sve “extra semi-volatile” parameters needed by HYSPLIT-SV run_name_sfg Simulation configuration parameters used by HYSPLIT-SV run_name_gem Simulation configuration parameters used by optional Global Eulerian Model (GEM) component of HYSPLIT-SV run_name_rfg Receptor setup configuration file, where area and point receptor groups are selected for inclusion in the simulation Mexico City Workshop, Feb 2012

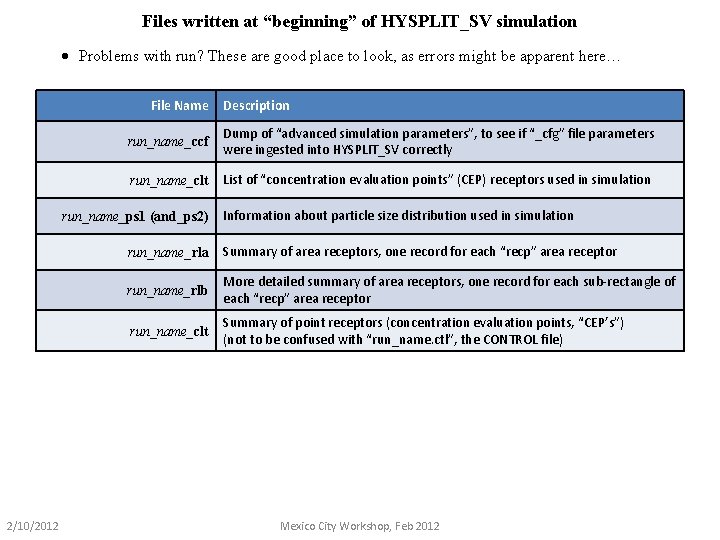
Files written at “beginning” of HYSPLIT_SV simulation · Problems with run? These are good place to look, as errors might be apparent here… File Name run_name_ccf Dump of “advanced simulation parameters”, to see if “_cfg” file parameters were ingested into HYSPLIT_SV correctly run_name_clt List of “concentration evaluation points” (CEP) receptors used in simulation run_name_ps 1 (and_ps 2) 2/10/2012 Description Information about particle size distribution used in simulation run_name_rla Summary of area receptors, one record for each “recp” area receptor run_name_rlb More detailed summary of area receptors, one record for each sub-rectangle of each “recp” area receptor run_name_clt Summary of point receptors (concentration evaluation points, “CEP’s”) (not to be confused with “run_name. ctl”, the CONTROL file) Mexico City Workshop, Feb 2012

“postscript image results” files produced after the HYSPLIT_SV simulation · These are produced using settings and choices established in the RUN and SET files; most of the settings are “hardwired” in the SET batch file but can be edited. · These files can get pretty big if the temporal_hrs setting is too small for long runs. File Name run_name_global. temporal. ps Postscript format deposition map over a global domain for PUFF results only, written as specified in the RUN and SET batch files, at the frequency specified by “temporal_hrs” in the SET batch file. This is produced by the program “gridplot”. Note that this program also produces ascii text output, as specified in SET batch file, for each output time step: run_name_global_temporal_PUF_001. txt, run_name_global_temporal_PUF_002. txt, … run_name_global. totrun. ps Postscript format deposition map over a global domain for PUFF results only, written as specified in the RUN and SET batch files, cumulative for the entire simulation. This is produced by the program “gridplot”, along with an ascii text output for each grid point: run_name_global_totrun_PUF_001. txt run_name_MEXICO. temporal. ps run_name_MEXICO. totrun. ps 2/10/2012 Description Postscript format deposition map over a MEXICO domain for PUFF results only, written as specified in RUN and SET batch files, at the frequency specified by “temporal_hrs” in the SET batch file. This is produced by the program “concplot”. Postscript format deposition map over a MEXICO domain for PUFF results only, written as specified in RUN and SET batch files, cumulative for the entire simulation. This is produced by the program “concplot”. Mexico City Workshop, Feb 2012

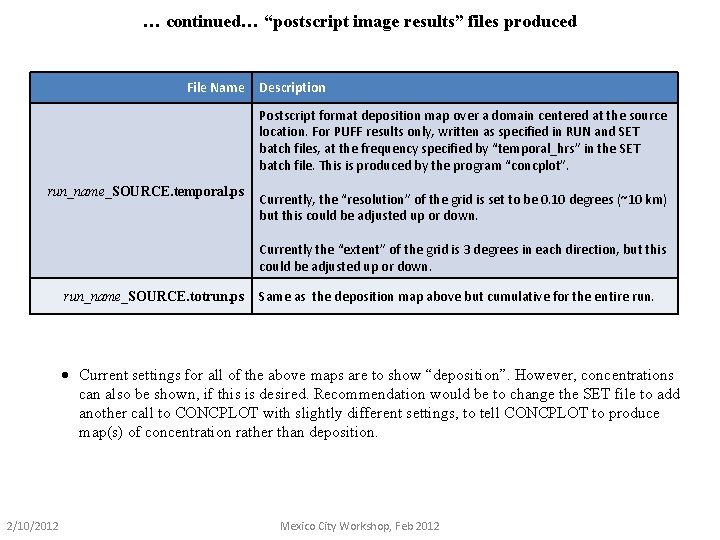
… continued… “postscript image results” files produced File Name Description Postscript format deposition map over a domain centered at the source location. For PUFF results only, written as specified in RUN and SET batch files, at the frequency specified by “temporal_hrs” in the SET batch file. This is produced by the program “concplot”. run_name_SOURCE. temporal. ps Currently, the “resolution” of the grid is set to be 0. 10 degrees (~10 km) but this could be adjusted up or down. Currently the “extent” of the grid is 3 degrees in each direction, but this could be adjusted up or down. run_name_SOURCE. totrun. ps Same as the deposition map above but cumulative for the entire run. · Current settings for all of the above maps are to show “deposition”. However, concentrations can also be shown, if this is desired. Recommendation would be to change the SET file to add another call to CONCPLOT with slightly different settings, to tell CONCPLOT to produce map(s) of concentration rather than deposition. 2/10/2012 Mexico City Workshop, Feb 2012

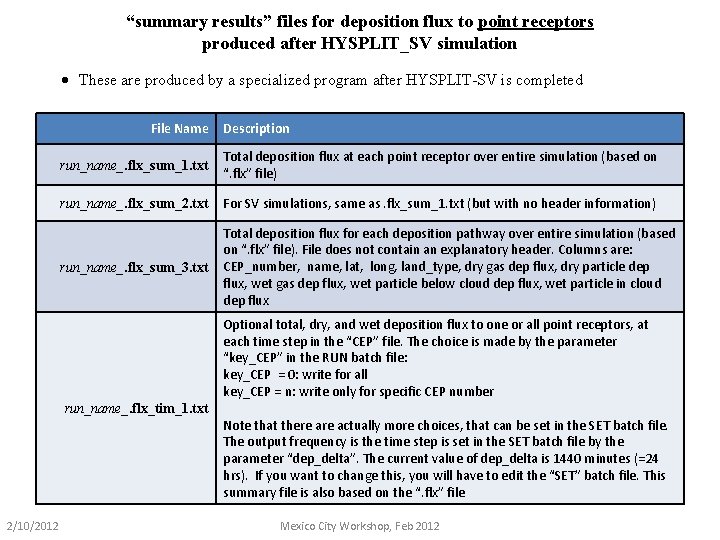
“summary results” files for deposition flux to point receptors produced after HYSPLIT_SV simulation · These are produced by a specialized program after HYSPLIT-SV is completed File Name Description run_name_. flx_sum_1. txt Total deposition flux at each point receptor over entire simulation (based on “. flx” file) run_name_. flx_sum_2. txt For SV simulations, same as. flx_sum_1. txt (but with no header information) run_name_. flx_sum_3. txt Total deposition flux for each deposition pathway over entire simulation (based on “. flx” file). File does not contain an explanatory header. Columns are: CEP_number, name, lat, long, land_type, dry gas dep flux, dry particle dep flux, wet gas dep flux, wet particle below cloud dep flux, wet particle in cloud dep flux Optional total, dry, and wet deposition flux to one or all point receptors, at each time step in the “CEP” file. The choice is made by the parameter “key_CEP” in the RUN batch file: key_CEP = 0: write for all key_CEP = n: write only for specific CEP number run_name_. flx_tim_1. txt Note that there actually more choices, that can be set in the SET batch file. The output frequency is the time step is set in the SET batch file by the parameter “dep_delta”. The current value of dep_delta is 1440 minutes (=24 hrs). If you want to change this, you will have to edit the “SET” batch file. This summary file is also based on the “. flx” file 2/10/2012 Mexico City Workshop, Feb 2012

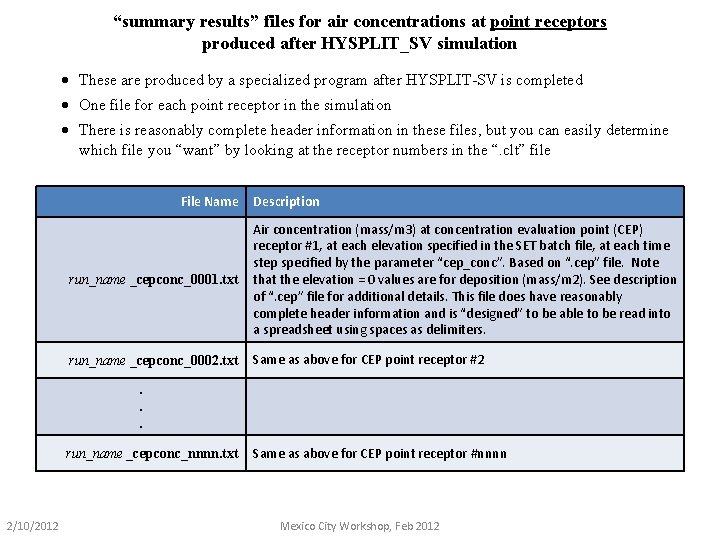
“summary results” files for air concentrations at point receptors produced after HYSPLIT_SV simulation · These are produced by a specialized program after HYSPLIT-SV is completed · One file for each point receptor in the simulation · There is reasonably complete header information in these files, but you can easily determine which file you “want” by looking at the receptor numbers in the “. clt” file File Name Description run_name _cepconc_0001. txt Air concentration (mass/m 3) at concentration evaluation point (CEP) receptor #1, at each elevation specified in the SET batch file, at each time step specified by the parameter “cep_conc”. Based on “. cep” file. Note that the elevation = 0 values are for deposition (mass/m 2). See description of “. cep” file for additional details. This file does have reasonably complete header information and is “designed” to be able to be read into a spreadsheet using spaces as delimiters. run_name _cepconc_0002. txt Same as above for CEP point receptor #2 . . . run_name _cepconc_nnnn. txt 2/10/2012 Same as above for CEP point receptor #nnnn Mexico City Workshop, Feb 2012

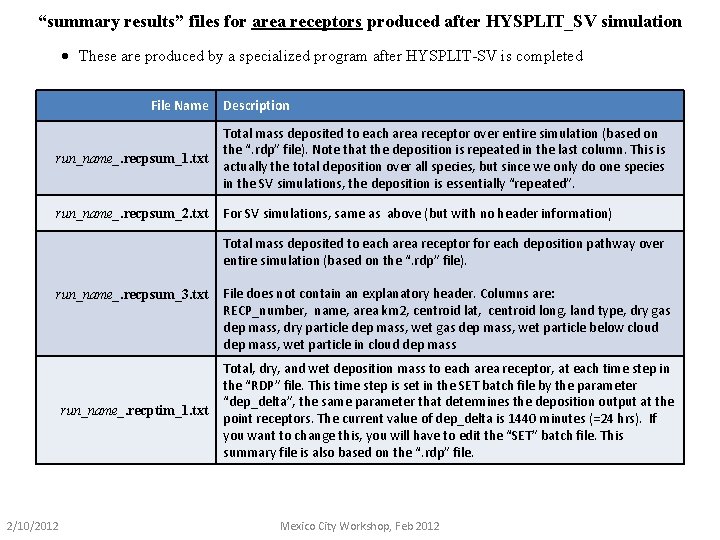
“summary results” files for area receptors produced after HYSPLIT_SV simulation · These are produced by a specialized program after HYSPLIT-SV is completed File Name Description run_name_. recpsum_1. txt Total mass deposited to each area receptor over entire simulation (based on the “. rdp” file). Note that the deposition is repeated in the last column. This is actually the total deposition over all species, but since we only do one species in the SV simulations, the deposition is essentially “repeated”. run_name_. recpsum_2. txt For SV simulations, same as above (but with no header information) Total mass deposited to each area receptor for each deposition pathway over entire simulation (based on the “. rdp” file). run_name_. recpsum_3. txt run_name_. recptim_1. txt 2/10/2012 File does not contain an explanatory header. Columns are: RECP_number, name, area km 2, centroid lat, centroid long, land type, dry gas dep mass, dry particle dep mass, wet gas dep mass, wet particle below cloud dep mass, wet particle in cloud dep mass Total, dry, and wet deposition mass to each area receptor, at each time step in the “RDP” file. This time step is set in the SET batch file by the parameter “dep_delta”, the same parameter that determines the deposition output at the point receptors. The current value of dep_delta is 1440 minutes (=24 hrs). If you want to change this, you will have to edit the “SET” batch file. This summary file is also based on the “. rdp” file. Mexico City Workshop, Feb 2012

“raw results” files produced during HYSPLIT_SV simulation File Name run_name_cep 2/10/2012 Description Concentration at each point receptor, at each elevation, at each “cepconc_delta” interval, as defined in the SET batch file. This is set at 180 minutes = 3 hours for initial testing. The elevations are also established in the SET batch file. Currently they are set at 0, 100, 500, 1000, 2000, 3000, and 4000 meters above ground level. If you want to change the elevations, you can. The “ 0” level is actually deposition. Units in this file are “mass/m 3” for concentrations and “mass/m 2” for deposition. Depending on the number of point receptors, the number of elevations, the frequency of output, and the run duration, this file can get pretty big. run_name_flx Deposition flux at each point receptor, for each deposition pathway, at each “dep_delta” interval, as defined in the SET batch file. This is set at 1440 minutes = 24 hours for initial testing. Units in this file are “mass/m 2”. run_name_rdp Deposition flux at each area receptor, for each deposition pathway, at each “dep_delta” interval, as defined in the SET batch file. This is set at 1440 minutes = 24 hours for initial testing. Units in this file are “mass”. Mexico City Workshop, Feb 2012

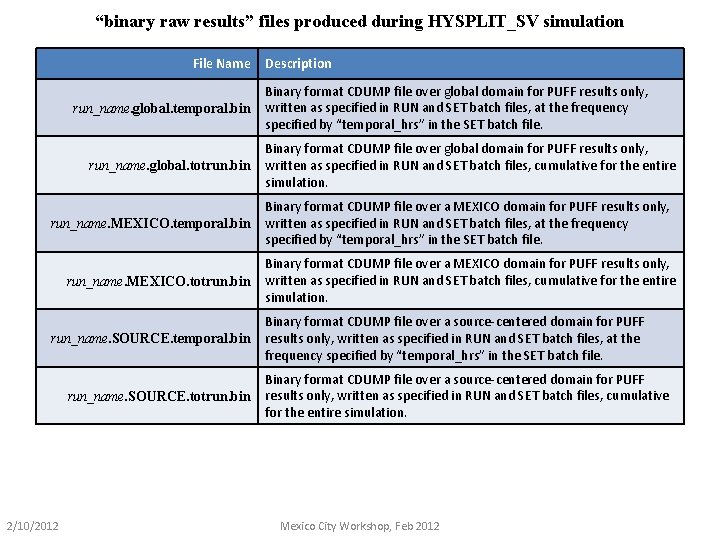
“binary raw results” files produced during HYSPLIT_SV simulation File Name run_name. global. temporal. bin run_name. global. totrun. bin run_name. MEXICO. temporal. bin run_name. MEXICO. totrun. bin run_name. SOURCE. temporal. bin run_name. SOURCE. totrun. bin 2/10/2012 Description Binary format CDUMP file over global domain for PUFF results only, written as specified in RUN and SET batch files, at the frequency specified by “temporal_hrs” in the SET batch file. Binary format CDUMP file over global domain for PUFF results only, written as specified in RUN and SET batch files, cumulative for the entire simulation. Binary format CDUMP file over a MEXICO domain for PUFF results only, written as specified in RUN and SET batch files, at the frequency specified by “temporal_hrs” in the SET batch file. Binary format CDUMP file over a MEXICO domain for PUFF results only, written as specified in RUN and SET batch files, cumulative for the entire simulation. Binary format CDUMP file over a source-centered domain for PUFF results only, written as specified in RUN and SET batch files, at the frequency specified by “temporal_hrs” in the SET batch file. Binary format CDUMP file over a source-centered domain for PUFF results only, written as specified in RUN and SET batch files, cumulative for the entire simulation. Mexico City Workshop, Feb 2012

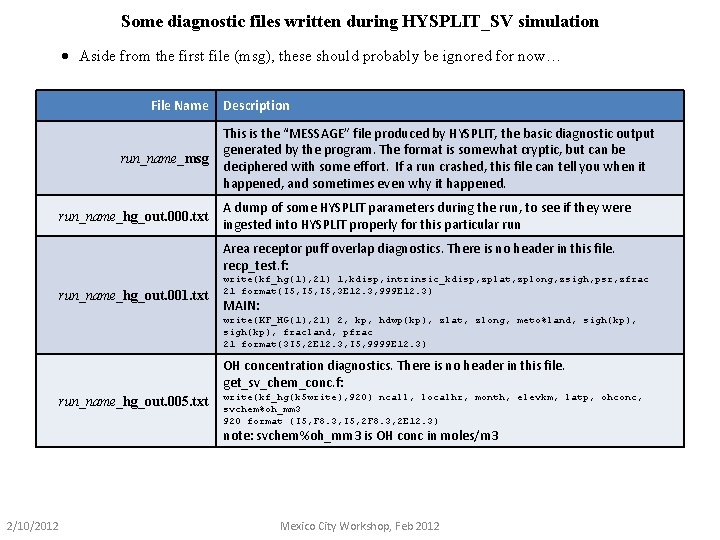
Some diagnostic files written during HYSPLIT_SV simulation · Aside from the first file (msg), these should probably be ignored for now… File Name run_name_msg run_name_hg_out. 000. txt Description This is the “MESSAGE” file produced by HYSPLIT, the basic diagnostic output generated by the program. The format is somewhat cryptic, but can be deciphered with some effort. If a run crashed, this file can tell you when it happened, and sometimes even why it happened. A dump of some HYSPLIT parameters during the run, to see if they were ingested into HYSPLIT properly for this particular run Area receptor puff overlap diagnostics. There is no header in this file. recp_test. f: run_name_hg_out. 001. txt write(kf_hg(1), 21) 1, kdisp, intrinsic_kdisp, zplat, zplong, zsigh, psr, zfrac 21 format(I 5, 3 E 12. 3, 999 E 12. 3) MAIN: write(KF_HG(1), 21) 2, kp, hdwp(kp), zlat, zlong, meto%land, sigh(kp), fracland, pfrac 21 format(3 I 5, 2 E 12. 3, I 5, 9999 E 12. 3) OH concentration diagnostics. There is no header in this file. get_sv_chem_conc. f: run_name_hg_out. 005. txt write(kf_hg(k 5 write), 920) ncall, localhr, month, elevkm, latp, ohconc, svchem%oh_mm 3 920 format (I 5, F 8. 3, I 5, 2 F 8. 3, 2 E 12. 3) note: svchem%oh_mm 3 is OH conc in moles/m 3 2/10/2012 Mexico City Workshop, Feb 2012

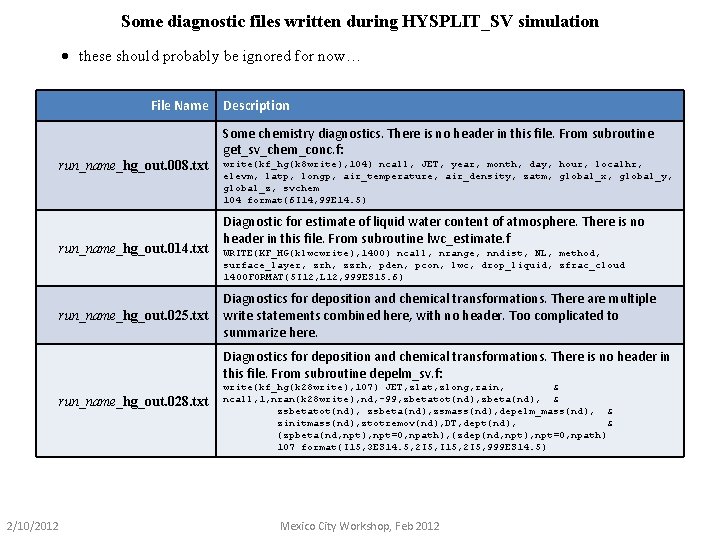
Some diagnostic files written during HYSPLIT_SV simulation · these should probably be ignored for now… File Name run_name_hg_out. 008. txt run_name_hg_out. 014. txt run_name_hg_out. 025. txt Description Some chemistry diagnostics. There is no header in this file. From subroutine get_sv_chem_conc. f: write(kf_hg(k 8 write), 104) ncall, JET, year, month, day, hour, localhr, elevm, latp, longp, air_temperature, air_density, zatm, global_x, global_y, global_z, svchem 104 format(6 I 14, 99 E 14. 5) Diagnostic for estimate of liquid water content of atmosphere. There is no header in this file. From subroutine lwc_estimate. f WRITE(KF_HG(klwcwrite), 1400) ncall, nrange, nndist, NL, method, surface_layer, zrh, zzrh, pden, pcon, lwc, drop_liquid, zfrac_cloud 1400 FORMAT(5 I 12, L 12, 999 ES 15. 6) Diagnostics for deposition and chemical transformations. There are multiple write statements combined here, with no header. Too complicated to summarize here. Diagnostics for deposition and chemical transformations. There is no header in this file. From subroutine depelm_sv. f: run_name_hg_out. 028. txt 2/10/2012 write(kf_hg(k 28 write), 107) JET, zlat, zlong, rain, & ncall, 1, nran(k 28 write), nd, -99, zbetatot(nd), zbeta(nd), & zsbetatot(nd), zsbeta(nd), zsmass(nd), depelm_mass(nd), & zinitmass(nd), ztotremov(nd), DT, dept(nd), & (zpbeta(nd, npt), npt=0, npath), (zdep(nd, npt), npt=0, npath) 107 format(I 15, 3 ES 14. 5, 2 I 5, I 15, 2 I 5, 999 ES 14. 5) Mexico City Workshop, Feb 2012

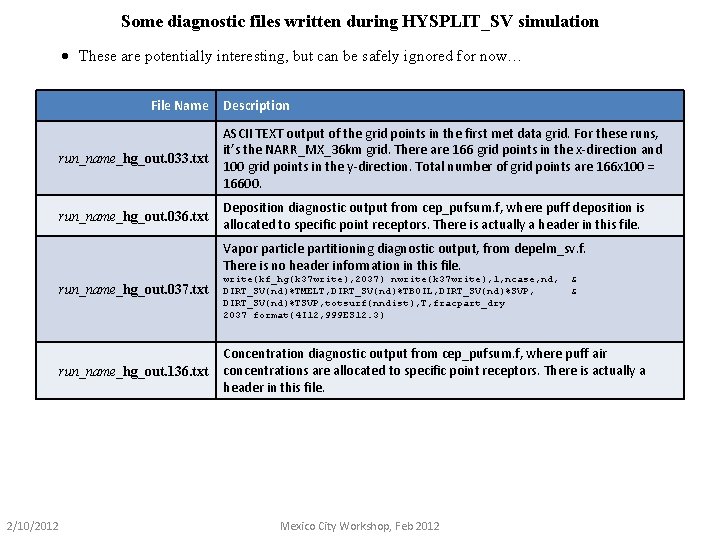
Some diagnostic files written during HYSPLIT_SV simulation · These are potentially interesting, but can be safely ignored for now… File Name Description run_name_hg_out. 033. txt ASCII TEXT output of the grid points in the first met data grid. For these runs, it’s the NARR_MX_36 km grid. There are 166 grid points in the x-direction and 100 grid points in the y-direction. Total number of grid points are 166 x 100 = 16600. run_name_hg_out. 036. txt Deposition diagnostic output from cep_pufsum. f, where puff deposition is allocated to specific point receptors. There is actually a header in this file. Vapor particle partitioning diagnostic output, from depelm_sv. f. There is no header information in this file. run_name_hg_out. 037. txt run_name_hg_out. 136. txt 2/10/2012 write(kf_hg(k 37 write), 2037) nwrite(k 37 write), 1, ncase, nd, DIRT_SV(nd)%TMELT, DIRT_SV(nd)%TBOIL, DIRT_SV(nd)%SVP, DIRT_SV(nd)%TSVP, totsurf(nndist), T, fracpart_dry 2037 format(4 I 12, 999 ES 12. 3) & & Concentration diagnostic output from cep_pufsum. f, where puff air concentrations are allocated to specific point receptors. There is actually a header in this file. Mexico City Workshop, Feb 2012

Some diagnostic files written during HYSPLIT_SV simulation · This is potentially interesting, but would need some effort to interpret and/or /utilize it… File Name Description Diagnostic output for chemical transformation, written each hour. This is the total chemical transformation mass of the pollutant – i. e. , the “destruction” of the pollutant by chemical reaction or photolysis – during the preceding hour. Imagine there is a total of X grams of mass in the model at the start of the hour. This file contains the model-estimated values of the total Y grams of mass in the model that were “destroyed” during the next hour. Thus, if there were no deposition or no loss of pollutant out the top or sides of the grid, the total mass of pollutant at the end of the hour would be X – Y. For pure PUF runs (without GEM) the GEM component will be zero. run_name_zdest_out. txt Here is the write statement in sv_chem. f write(2060, 2061) JET, kyear, kmonth, kday, khour, kminute, & (zdest_puf(nd), nd=1, numpol), (zdest_gem(nd), nd=1, numpol), & (zdest_puf(nd)+zdest_gem(nd), nd=1, numpol) 2061 format(I 15, 5 I 5, 9999 E 15. 6) Note that in the above write statement, numpol=1 for these HYSPLIT 4_SV runs, i. e. , there is only one species per run. So, the columns in the file are simply: JET, year, month, hour, minute, zdest_puf, zdest_gem, zdest_tot Where JET = “J elapsed time”, an internal time stamp in HYSPLIT, referring to the cumulative elapsed minutes since the beginning of 1970. And where zdest_tot = zdest_puf + zdest_gem 2/10/2012 Mexico City Workshop, Feb 2012

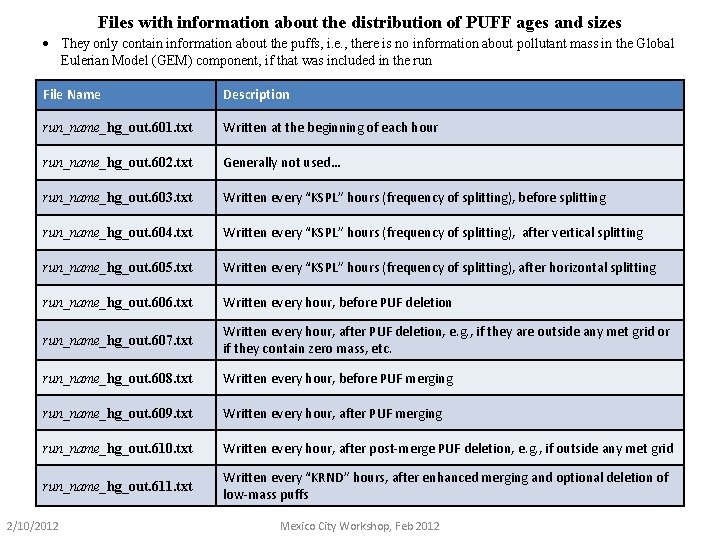
Files with information about the distribution of PUFF ages and sizes · They only contain information about the puffs, i. e. , there is no information about pollutant mass in the Global Eulerian Model (GEM) component, if that was included in the run File Name Description run_name_hg_out. 601. txt Written at the beginning of each hour run_name_hg_out. 602. txt Generally not used… run_name_hg_out. 603. txt Written every “KSPL” hours (frequency of splitting), before splitting run_name_hg_out. 604. txt Written every “KSPL” hours (frequency of splitting), after vertical splitting run_name_hg_out. 605. txt Written every “KSPL” hours (frequency of splitting), after horizontal splitting run_name_hg_out. 606. txt Written every hour, before PUF deletion run_name_hg_out. 607. txt Written every hour, after PUF deletion, e. g. , if they are outside any met grid or if they contain zero mass, etc. run_name_hg_out. 608. txt Written every hour, before PUF merging run_name_hg_out. 609. txt Written every hour, after PUF merging run_name_hg_out. 610. txt Written every hour, after post-merge PUF deletion, e. g. , if outside any met grid run_name_hg_out. 611. txt Written every “KRND” hours, after enhanced merging and optional deletion of low-mass puffs 2/10/2012 Mexico City Workshop, Feb 2012

A SET OF INITIAL TEST RUNS FOR: 2, 3, 7, 8 -TCDD 2, 3, 4, 7, 8 -Pe. CDF 2, 3, 7, 8 -TCDF and OCDD 2/10/2012 Mexico City Workshop, Feb 2012

If desired, several other test runs can be done. DOS Batch files for these test runs are also provided in the working_SV directory. These runs have also been “pre-computed”, and all the results are available at the FTP site under the folder: “ADDITIONAL_pre_computed_test_runs”. A set of 168 -hr runs from std pts 1 thru 5, with each batch file containing separate runs for 2, 3, 7, 8 -TCDF, 2, 3, 7, 8 TCDD, 2, 3, 4, 7, 8 -Pe. CDF, and OCDD, with a time step of 60 minutes: • • • run_stdpt_1_168 hr. bat run_stdpt_2_168 hr. bat run_stdpt_3_168 hr. bat run_stdpt_4_168 hr. bat run_stdpt_5_168 hr. bat A set of 672 -hr runs from std pts 1 thru 5, with each batch file containing separate runs for 2, 3, 7, 8 -TCDF, 2, 3, 7, 8 TCDD, 2, 3, 4, 7, 8 -Pe. CDF, and OCDD, with a time step of 30 minutes: • • • run_stdpt_1_672 hr. bat run_stdpt_2_672 hr. bat run_stdpt_3_672 hr. bat run_stdpt_4_672 hr. bat run_stdpt_5_672 hr. bat A set of runs showing an example of parameter variation, a process that should be explored further as the project proceeds. In this case a series of 672 -hr runs from std pt 2 for 2, 3, 7, 8 -TCDD, for different values of “maxpar”, the maximum number of puffs allowed in the simulation at any one time. In all the simulations above, a value of 10, 000 was used for “maxpar”. Here a series of runs is done with values of 1000, 2000, 5000, 20000, and 50000, to compare with the run done earlier with maxpar=10000 • run_stdpt_2_tcdd_672 hr_maxpar. bat 2/10/2012 Mexico City Workshop, Feb 2012

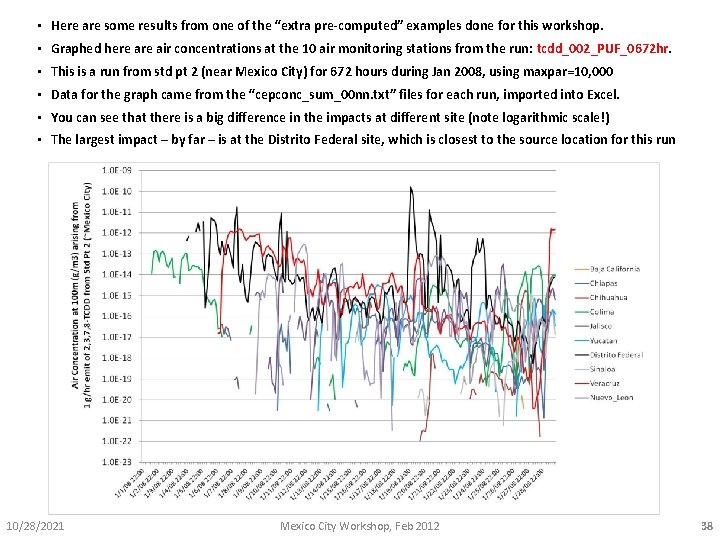
• Here are some results from one of the “extra pre-computed” examples done for this workshop. • Graphed here air concentrations at the 10 air monitoring stations from the run: tcdd_002_PUF_0672 hr. • This is a run from std pt 2 (near Mexico City) for 672 hours during Jan 2008, using maxpar=10, 000 • Data for the graph came from the “cepconc_sum_00 nn. txt” files for each run, imported into Excel. • You can see that there is a big difference in the impacts at different site (note logarithmic scale!) • The largest impact – by far – is at the Distrito Federal site, which is closest to the source location for this run 10/28/2021 Mexico City Workshop, Feb 2012 38

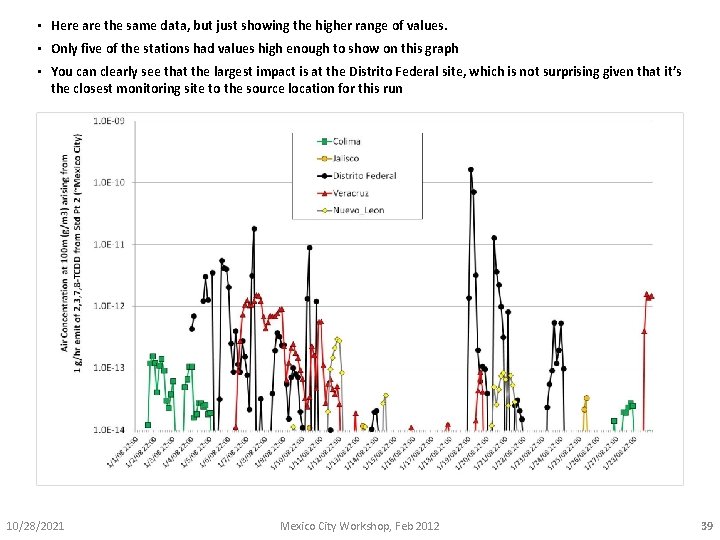
• Here are the same data, but just showing the higher range of values. • Only five of the stations had values high enough to show on this graph • You can clearly see that the largest impact is at the Distrito Federal site, which is not surprising given that it’s the closest monitoring site to the source location for this run 10/28/2021 Mexico City Workshop, Feb 2012 39

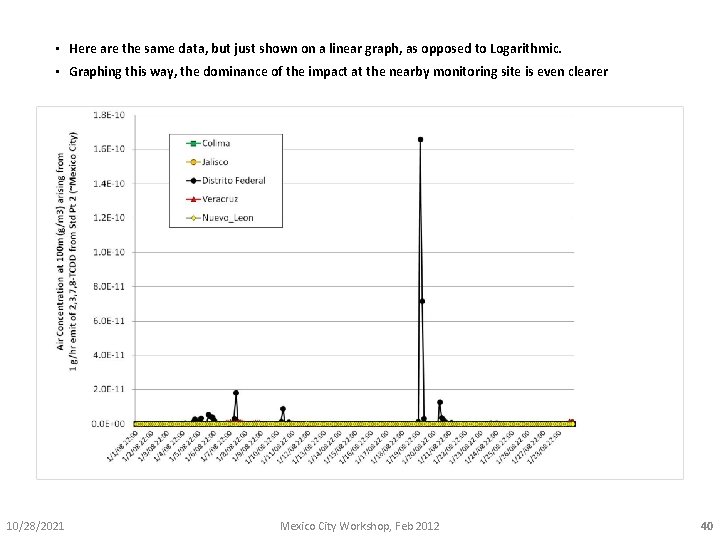
• Here are the same data, but just shown on a linear graph, as opposed to Logarithmic. • Graphing this way, the dominance of the impact at the nearby monitoring site is even clearer 10/28/2021 Mexico City Workshop, Feb 2012 40

SOME KEY PARAMETERS to CONSIDER 2/10/2012 Mexico City Workshop, Feb 2012

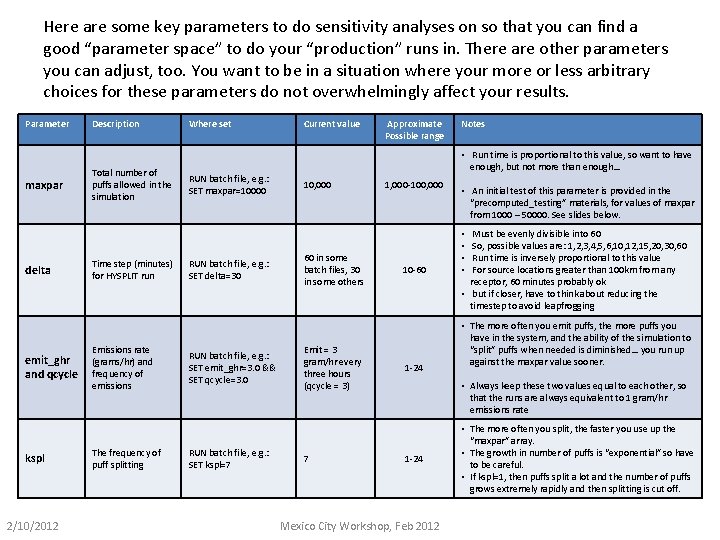
Here are some key parameters to do sensitivity analyses on so that you can find a good “parameter space” to do your “production” runs in. There are other parameters you can adjust, too. You want to be in a situation where your more or less arbitrary choices for these parameters do not overwhelmingly affect your results. Parameter maxpar delta emit_ghr and qcycle kspl 2/10/2012 Description Total number of puffs allowed in the simulation Time step (minutes) for HYSPLIT run Emissions rate (grams/hr) and frequency of emissions The frequency of puff splitting Where set RUN batch file, e. g. : SET maxpar=10000 RUN batch file, e. g. : SET delta=30 RUN batch file, e. g. : SET emit_ghr=3. 0 && SET qcycle=3. 0 RUN batch file, e. g. : SET kspl=7 Current value Approximate Possible range Notes • Run time is proportional to this value, so want to have enough, but not more than enough… 10, 000 60 in some batch files, 30 in some others Emit = 3 gram/hr every three hours (qcycle = 3) 7 1, 000 -100, 000 10 -60 1 -24 • An initial test of this parameter is provided in the “precomputed_testing” materials, for values of maxpar from 1000 – 50000. See slides below. Must be evenly divisible into 60 So, possible values are: 1, 2, 3, 4, 5, 6, 10, 12, 15, 20, 30, 60 Run time is inversely proportional to this value For source locations greater than 100 km from any receptor, 60 minutes probably ok • but if closer, have to think about reducing the timestep to avoid leapfrogging • • • The more often you emit puffs, the more puffs you have in the system, and the ability of the simulation to “split” puffs when needed is diminished… you run up against the maxpar value sooner. • Always keep these two values equal to each other, so that the runs are always equivalent to 1 gram/hr emissions rate 1 -24 Mexico City Workshop, Feb 2012 • The more often you split, the faster you use up the “maxpar” array. • The growth in number of puffs is “exponential” so have to be careful. • If kspl=1, then puffs split a lot and the number of puffs grows extremely rapidly and then splitting is cut off.

A example of a sensitivity analysis, examining the effect of MAXPAR on the deposition and air concentrations estimated by the model 2/10/2012 Mexico City Workshop, Feb 2012

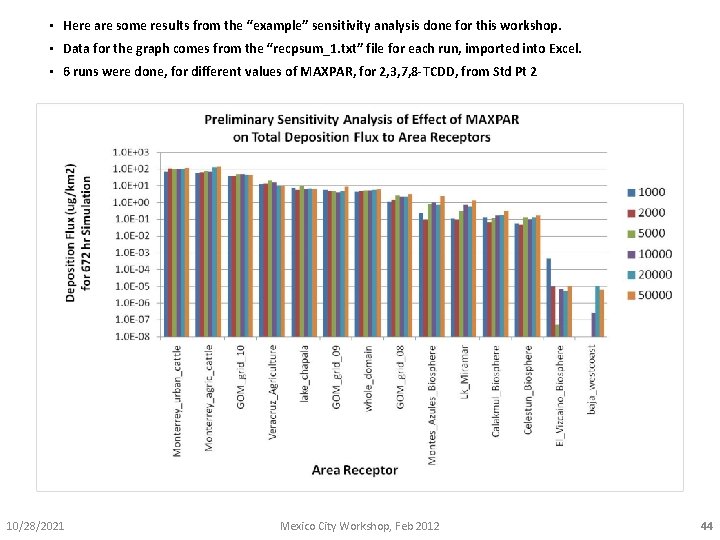
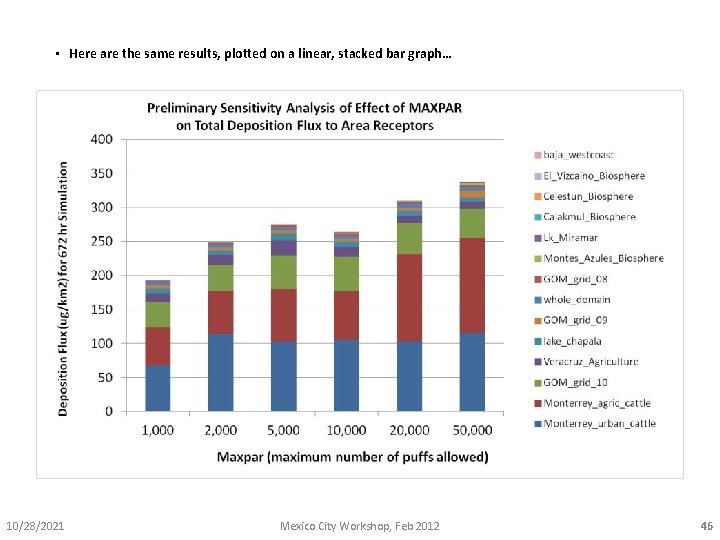
• Here are some results from the “example” sensitivity analysis done for this workshop. • Data for the graph comes from the “recpsum_1. txt” file for each run, imported into Excel. • 6 runs were done, for different values of MAXPAR, for 2, 3, 7, 8 -TCDD, from Std Pt 2 10/28/2021 Mexico City Workshop, Feb 2012 44

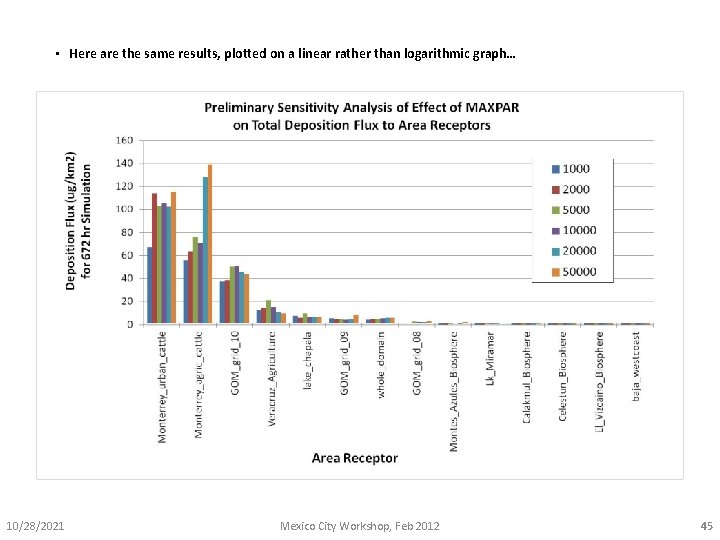
• Here are the same results, plotted on a linear rather than logarithmic graph… 10/28/2021 Mexico City Workshop, Feb 2012 45

• Here are the same results, plotted on a linear, stacked bar graph… 10/28/2021 Mexico City Workshop, Feb 2012 46

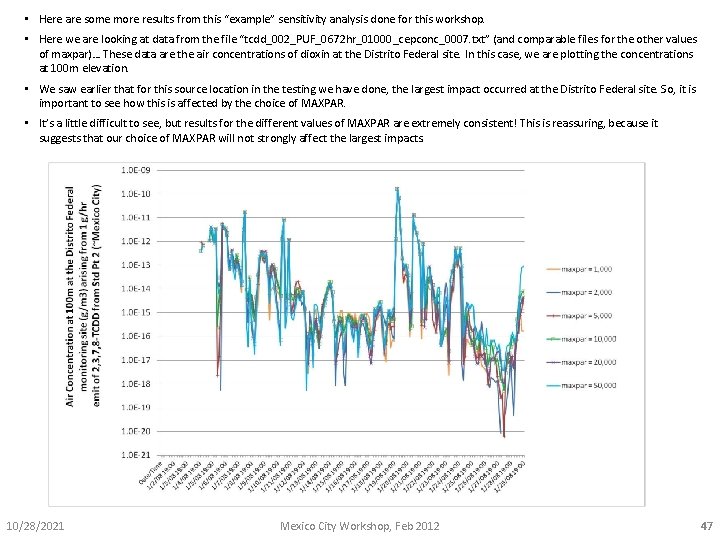
• Here are some more results from this “example” sensitivity analysis done for this workshop. • Here we are looking at data from the file “tcdd_002_PUF_0672 hr_01000_cepconc_0007. txt” (and comparable files for the other values of maxpar)… These data are the air concentrations of dioxin at the Distrito Federal site. In this case, we are plotting the concentrations at 100 m elevation. • We saw earlier that for this source location in the testing we have done, the largest impact occurred at the Distrito Federal site. So, it is important to see how this is affected by the choice of MAXPAR. • It’s a little difficult to see, but results for the different values of MAXPAR are extremely consistent! This is reassuring, because it suggests that our choice of MAXPAR will not strongly affect the largest impacts. 10/28/2021 Mexico City Workshop, Feb 2012 47

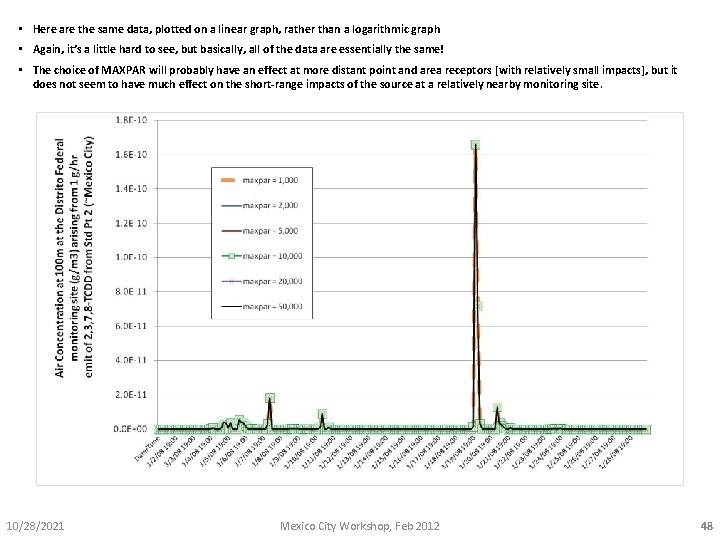
• Here are the same data, plotted on a linear graph, rather than a logarithmic graph • Again, it’s a little hard to see, but basically, all of the data are essentially the same! • The choice of MAXPAR will probably have an effect at more distant point and area receptors [with relatively small impacts], but it does not seem to have much effect on the short-range impacts of the source at a relatively nearby monitoring site. 10/28/2021 Mexico City Workshop, Feb 2012 48

What do these initial MAXPAR sensitivity analyses results suggest? • For receptors with significant impacts (higher deposition fluxes and/or higher air concentrations) MAXPAR does not make a huge difference. • This test was only done for one standard source location, for one species, and only for 672 hours. It would probably be useful to repeat this test for other locations and for longer time periods. • For receptors with “trace” impacts, MAXPAR will probably make a bigger difference, but it doesn’t matter very much, since the impacts are so small • A value of MAXPAR of 10, 000 -20, 000 will probably sufficient, but this should be confirmed with additional sensitivity analyses, including longer term simulations • The impact of MAXPAR is influenced by other settings. The settings for QCYCLE and KSPL also affect the number of puffs… So, if you find a good value for MAXPAR for a given set of QCYCLE/KSPL settings, this may not be a good values for other QCYCLE or KSPL settings. • It is recommended that you just use the NARR-MX-36 km grid, and not include the global met grid. This is because you don’t necessarily want to spend computational resources on puffs that are far away from Mexico. If you do want to simulate puffs on a global grid, you’d probably need to have a larger value of MAXPAR • The run time is directly proportional to MAXPAR… so, you want to have enough to get a decent simulation, but not so many that the runs take too long… 10/28/2021 Mexico City Workshop, Feb 2012 49

A possible outline of some of the next steps 2/10/2012 Mexico City Workshop, Feb 2012

1. Gain additional experience with the batch files and running the model 2. Carry out some sensitivity analyses on key parameters. 3. Gain experience with what the different output files are, and what they are telling us. 4. At some point, start doing longer runs, comparable to what you might be doing for the full analysis, e. g. , 13 month runs starting Dec 1, 2007 (one month spin-up to a full year 2008 simulation 5. Make “final” decisions on parameter choices and output choices, e. g. , keeping in mind issues of file size and run times. 6. Finalize specifications and choices of receptors. Once you start doing “production” runs, you want to keep all these constant. 7. Decide on a number of “standard source locations” or “standard points” to run. Key to this will be an examination of the inventory. We will want to run from areas of strong source intensity, especially if they are near the monitoring sites. We will also want to have a basic geographical “spread” of the standard points so that the spatial interpolation later will have a reasonable basis on which to operate. 8. Depending on choices, etc. , there may end up being from 20 – 200 standard points, plus or minus. 9. From each standard point, we will do 4 runs: one for “tcdf” (actually 2, 3, 7, 8 -TCDF), one for “tcdd” (actually 2, 3, 7, 8 -TCDD), one for “pcdf” (actually 2, 3, 4, 7, 8 -Pe. CDF) and one for “ocdd” (OCDD). These congeners span the range of fate/transport phenomena, and the impacts of the other emitted congeners can be estimated from the modeled impacts of these four. 10. It will be important to be careful about the time step during the runs. For standard source locations nearby monitoring sites or area receptors, smaller times steps may be required, to avoid “leapfrogging” … continued on next page… 10/28/2021 Mexico City Workshop, Feb 2012 51

11. In parallel with the above, continue to develop the congener-specific emissions inventory. 12. Ultimately, we need to be able to estimate for each source, the grams/hr of each congener and homologue group that is emitted. We also need accurate lat/long information for each source, as well as any other identifying characteristics that we want to carry through the modeling. 13. Once all the runs are done, and we have at least a preliminary inventory, we will then be able to run the “FINCALC” program to combine the unit-emission, four-congener runs with the inventory. The program uses spatial interpolation and congener interpolation to estimate the impact of each source on each receptor. The results will be given in terms of each congener and also for total PCDD/F and also for total TEQ, and we will have estimated deposition and air concentrations at each receptor that we have defined. 14. Note that Mark and Paul will be developing the FINCALC program for use in this project. Work on this has begun, but it will take some time before we have the program ready to be used for this project. However, the goal is to have the program ready at least by the time we are ready to use it, i. e. , when a certain number of runs have been done, and we have an emissions inventory that can be used. 15. Once we run FINCALC, we will be able to compare the model-estimated concentrations at each of the monitoring sites against the actual measurements. The degree of consistency between the two will be very instructive. If they are relatively consistent, this suggests that our inventory was very accurate and our modeling was very realistic. 16. In practice, the results are usually not “perfect” the first time. Often, the results of the analysis suggest, for example, that more standard source locations are needed, and/or that improvements in the emissions inventory are needed. 17. However, it is hoped and expected that ultimately, we will be able to get a reasonable match between the model predictions and the measurements. 18. At that point we will be able to confidently say that the results on deposition and source-attribution for that deposition to ecosystems throughout Mexico are relatively robust, and can be used to inform policy and additional research! 10/28/2021 Mexico City Workshop, Feb 2012 52