More on sourmash gather vs lca sourmash gather















- Slides: 25

More on sourmash
'gather’ vs ‘lca’

’sourmash gather’ – iterative, greedy approach on genomes.

lca = “lowest common ancestor”, assigns taxonomic identity to k-mers.

’sourmash lca gather’ – iterative, greedy approach on taxonomic lineages. We understand this approach less well than ‘sourmash gather’ : )


Synthetic metagenomic data https: //genomebiology. biomedcentral. com/articles/10. 1186/s 13059 -017 -1299 -7 Emerging datasets from the U. S. National Institute of Standards and Technology (NIST) for benchmarking metagenomic classifiers https: //ftp-private. ncbi. nlm. nih. gov/nist-immsa/IMMSA Mc. Intyre et al. , 2017. https: //www. nist. gov/mml/bbd/immsa-missionstatement

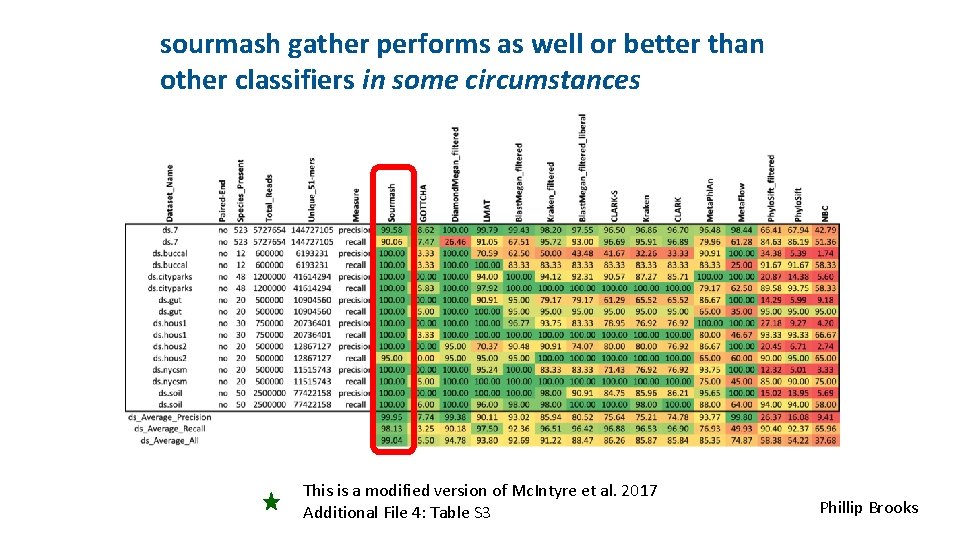
sourmash gather performs as well or better than other classifiers in some circumstances This is a modified version of Mc. Intyre et al. 2017 Additional File 4: Table S 3 Phillip Brooks

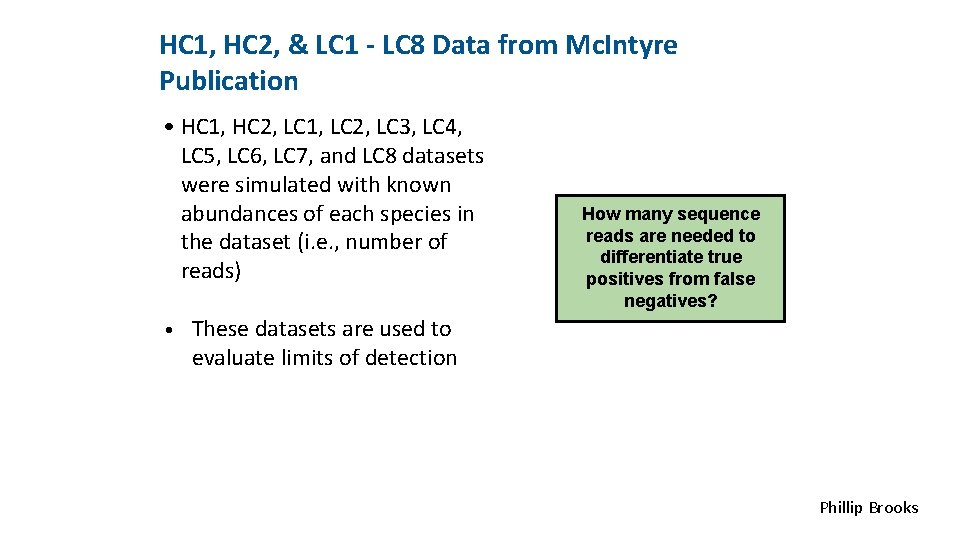
<INSERT CONTROL MARKING HERE> HC 1, HC 2, & LC 1 - LC 8 Data from Mc. Intyre Publication • HC 1, HC 2, LC 1, LC 2, LC 3, LC 4, LC 5, LC 6, LC 7, and LC 8 datasets were simulated with known abundances of each species in the dataset (i. e. , number of reads) How many sequence reads are needed to differentiate true positives from false negatives? • These datasets are used to evaluate limits of detection Phillip Brooks

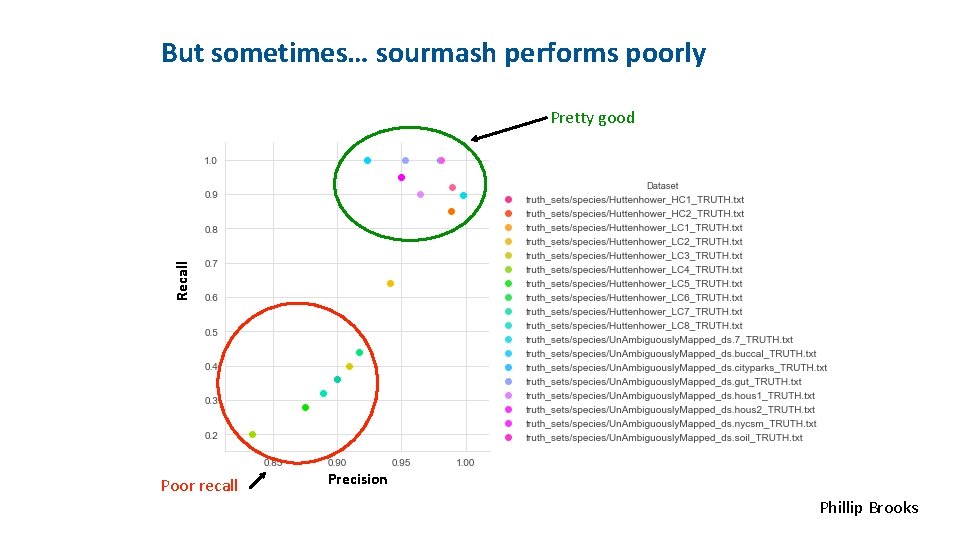
<INSERT CONTROL MARKING HERE> But sometimes… sourmash performs poorly Recall Pretty good Poor recall Precision Phillip Brooks

<INSERT CONTROL MARKING HERE> False Negatives Prevalent with <7, 000 reads Reads Per Species 30, 000 - 189, 000 20, 000 - 30, 000 16, 000 - 20, 000 15, 000 - 16, 000 14, 000 - 15, 000 13, 000 - 14, 000 12, 000 - 13, 000 11, 000 - 12, 000 10, 000 - 11, 000 9, 000 - 10, 000 8, 000 - 9, 000 7, 000 - 8, 000 6, 000 - 7, 000 5, 000 - 6, 000 4, 000 - 5, 000 3, 000 - 4, 000 2, 000 - 3, 000 1, 000 - 2, 000 0 - 1, 000 Correct Species 15 15 17 13 16 20 11 16 12 16 19 19 25 10 4 1 0 False Negatives 0 0 0 1 3 3 7 19 36 69 False negatives began appearing at <7, 000 reads per species. The exact number of reads and unique 51 -mers needed for a correct species call varies depending on the organism, reference database, and genomic regions covered by the reads. Phillip Brooks

<INSERT CONTROL MARKING HERE>

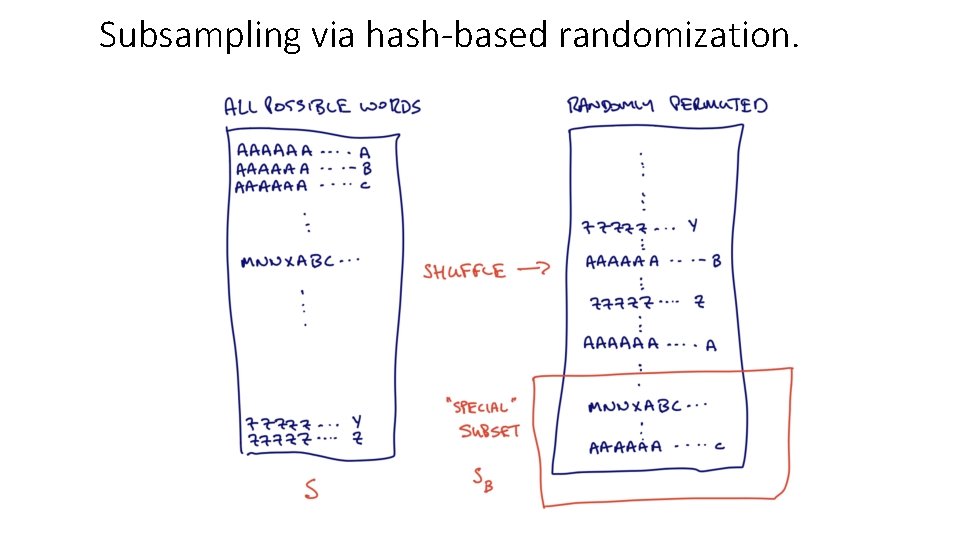
Subsampling via hash-based randomization.

Subsampling via hash-based randomization.

Subsampling via hash-based randomization.

This subsampling allows two estimators:

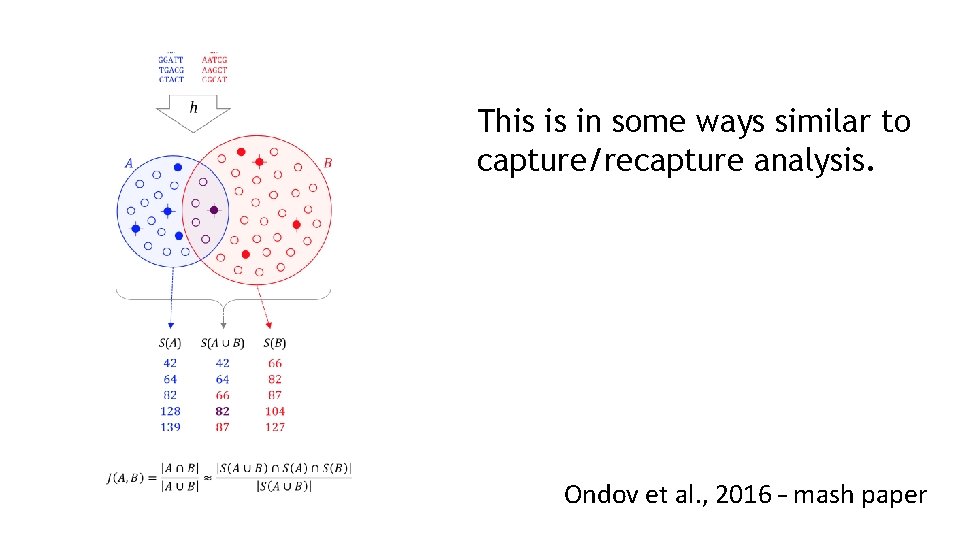
This is in some ways similar to capture/recapture analysis. Ondov et al. , 2016 – mash paper

Summary of approach: • Choose 1 / B words to be “special”, and subsample word collections down to those special words. • • If B = 1, you are picking all words! If B = 2, you are picking half the words! If B = 10, one tenth… Typically we choose B to be 1000, or 10, 000. • If “special” word is in a sample, it will always be in subsample (unlike Min. Hash). • Work only with these (much) smaller subsamples.

Features of approach: • Provides fast and lightweight estimation of similarity, containment; • B is tunable, with tradeoffs in resolution vs time & memory. • Subsampling to larger B is straightforward and does not involve revisiting raw data. • Incredibly computationally convenient! All of the analyses in this talk (over a Tbp of data) can be done on my laptop.

<INSERT CONTROL MARKING HERE>

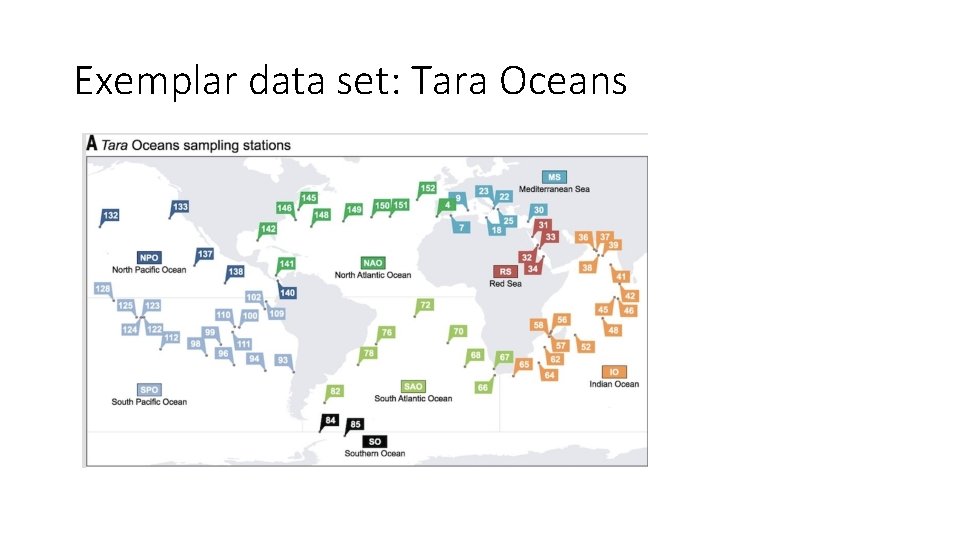
Exemplar data set: Tara Oceans Approximately 1 TB of data, in ~250 different samples (“libraries”). Sunagawa et al. , 2015

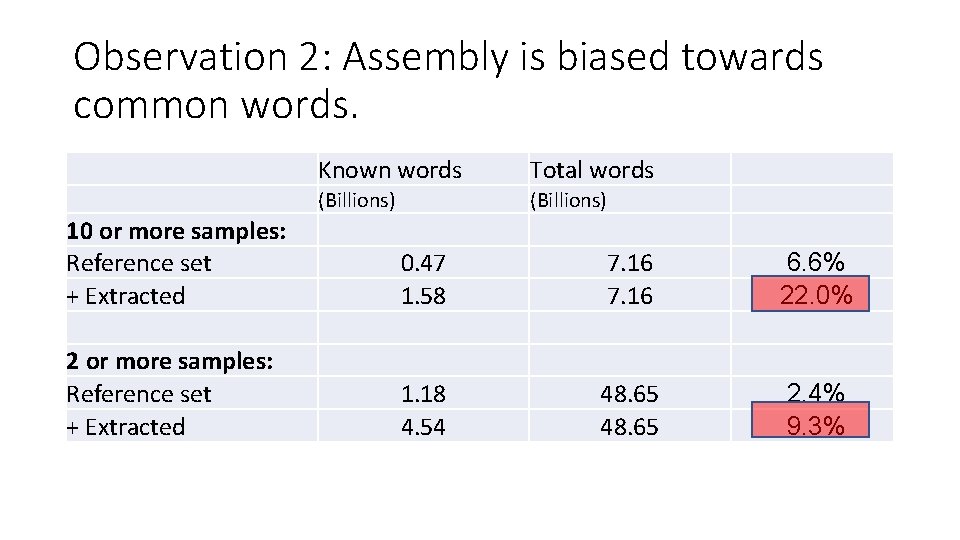
Tara vs genbank+JGI, and extracted via assembly: Known words Total words (Billions) 10 or more samples: Reference set + Extracted 0. 47 1. 58 7. 16 6. 6% 22. 0% 2 or more samples: Reference set + Extracted 1. 18 4. 54 48. 65 2. 4% 9. 3% Tara ocean data, k=31.

Observation 1: Vast majority is unknown. Known words Total words (Billions) 10 or more samples: Reference set + Extracted 0. 47 1. 58 7. 16 6. 6% 22. 0% 2 or more samples: Reference set + Extracted 1. 18 4. 54 48. 65 2. 4% 9. 3% Tara ocean data, k=31.

Observation 2: Assembly is biased towards common words. Known words Total words (Billions) 10 or more samples: Reference set + Extracted 0. 47 1. 58 7. 16 6. 6% 22. 0% 2 or more samples: Reference set + Extracted 1. 18 4. 54 48. 65 2. 4% 9. 3% Tara ocean data, k=31.

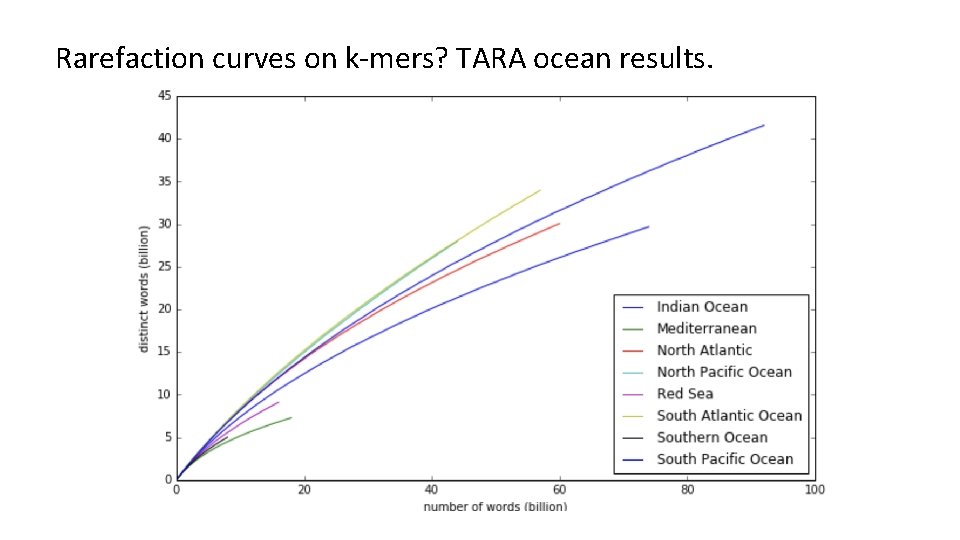
<INSERT CONTROL MARKING HERE> Rarefaction curves on k-mers? TARA ocean results.
 More more more i want more more more more we praise you
More more more i want more more more more we praise you More more more i want more more more more we praise you
More more more i want more more more more we praise you Lci
Lci Lca stata
Lca stata Hensriks
Hensriks Diagramme de flux lca
Diagramme de flux lca Lca analiza
Lca analiza Life cycle inventory
Life cycle inventory Lca payout cognizant
Lca payout cognizant Functional unit lca
Functional unit lca Lca
Lca Xilinx lca
Xilinx lca Glossaire lca cnci
Glossaire lca cnci Simapro lca software free download
Simapro lca software free download Career investigation task lca
Career investigation task lca Lca banrisul
Lca banrisul Contingent workforce compliance
Contingent workforce compliance Diagramme de flux lca
Diagramme de flux lca Randomisation en cluster
Randomisation en cluster Bees lca
Bees lca Lca lad
Lca lad Fonte immagine
Fonte immagine Distenze lca
Distenze lca The more you study the more you learn
The more you study the more you learn More love to thee o lord
More love to thee o lord Aspire not to have more but to be more
Aspire not to have more but to be more