Molecular Markers Definition Molecular marker is any site














![SSR - methodolgy Genotype A Genotype B [AT]1 8 [AT]2 2 [AT]1 [AT]2 PCR SSR - methodolgy Genotype A Genotype B [AT]1 8 [AT]2 2 [AT]1 [AT]2 PCR](https://slidetodoc.com/presentation_image_h2/41221deccec3d7ab92acdb34433742d0/image-24.jpg)


- Slides: 38

Molecular Markers

Definition: � “Molecular marker is any site (locus) in the genome of an organism at which the DNA base sequence varies among the different individuals of a population. A DNA sequence that is readily detected and whose inheritance can be easily monitered. ”


What are Molecular Markers: Specific fragments of DNA that can be identified within the whole genome. Molecular markers are the general assays that allow detection of the sequence differences between two or more individual. Molecular markers are found at specific locations of the genome. They are used to 'flag' the position of a particular gene or the inheritance of a particular characteristic or desired characteristics

� It can be used to reveal certain characteristics about the respective source. DNA, for example, is a molecular marker containing information about genetic disorders, genealogy and the evolutionary history of life. � Such markers generally have no apparent effect on the phenotype of the individual, but they can be determined by biochemical analysis of the DNA and are used for a variety of purposes, including chromosome mapping, DNA fingerprinting, and genetic screening.

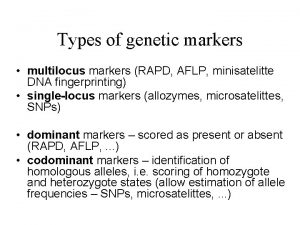
Types: There are two basic types of molecular markers: 1. Genetic Markers. 2. Biochemical Markers.

Genetic Markers: � A genetic marker is a gene or DNA sequence with a known location on a chromosome that can be used to identify individuals or species. It can be described as a variation (which may arise due to mutation or alteration in the genomic loci) that can be observed. � These are used to differentiate between individuals also in identifying DNA, DNA fingerprinting in crime investigations.

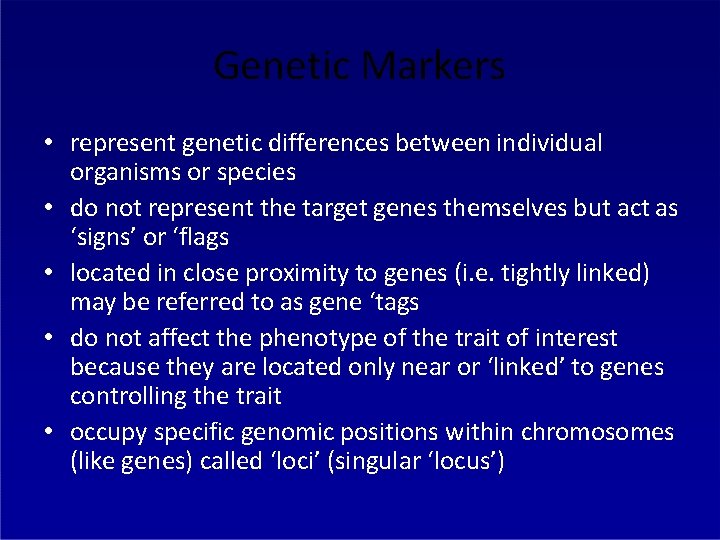
Genetic Markers • represent genetic differences between individual organisms or species • do not represent the target genes themselves but act as ‘signs’ or ‘flags • located in close proximity to genes (i. e. tightly linked) may be referred to as gene ‘tags • do not affect the phenotype of the trait of interest because they are located only near or ‘linked’ to genes controlling the trait • occupy specific genomic positions within chromosomes (like genes) called ‘loci’ (singular ‘locus’)

Classification of Genetic Markers • Classical markers • Morphological markers, • Cytological markers • Biochemical markers • DNA markers • RFLP, AFLP, RAPD, SSR, SNP

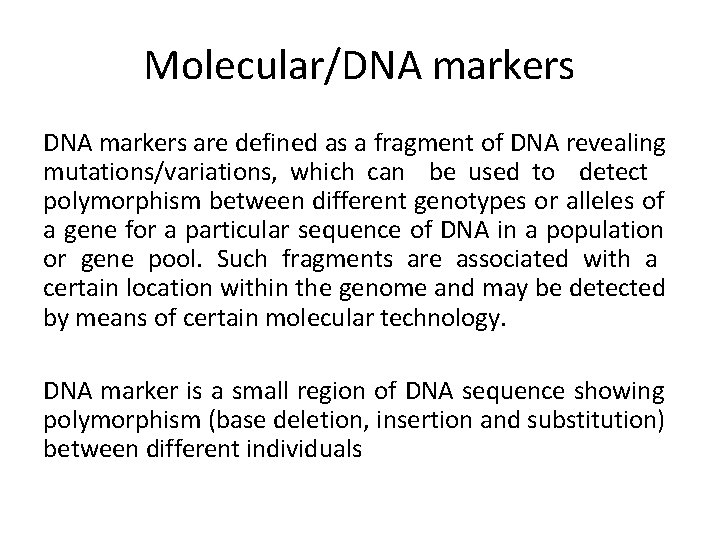
Molecular/DNA markers are defined as a fragment of DNA revealing mutations/variations, which can be used to detect polymorphism between different genotypes or alleles of a gene for a particular sequence of DNA in a population or gene pool. Such fragments are associated with a certain location within the genome and may be detected by means of certain molecular technology. DNA marker is a small region of DNA sequence showing polymorphism (base deletion, insertion and substitution) between different individuals

Criteria for ideal DNA markers • • • selectively neutral because they are usually located in non-coding regions of DNA High level of polymorphism unlimited in number and are not affected by environmental factors and/or the developmental stage of the plant Even distribution across the whole genome (not clustered in certain regions) Co-dominance in expression (so that heterozygotes can be distinguished from homozygotes) Clear distinct allelic features (so that the different alleles can be easily identified) Single copy and no pleiotropic effect Low cost to use (or cost-efficient marker development and genotyping) Easy assay/detection and automation High availability (un-restricted use) and suitability to be duplicated/multiplexed (so that the data can be accumulated and shared between laboratories) No detrimental effect on phenotype However, no molecular markers fulfill all these characteristics. Researchers choose the molecular marker according to their need and availability



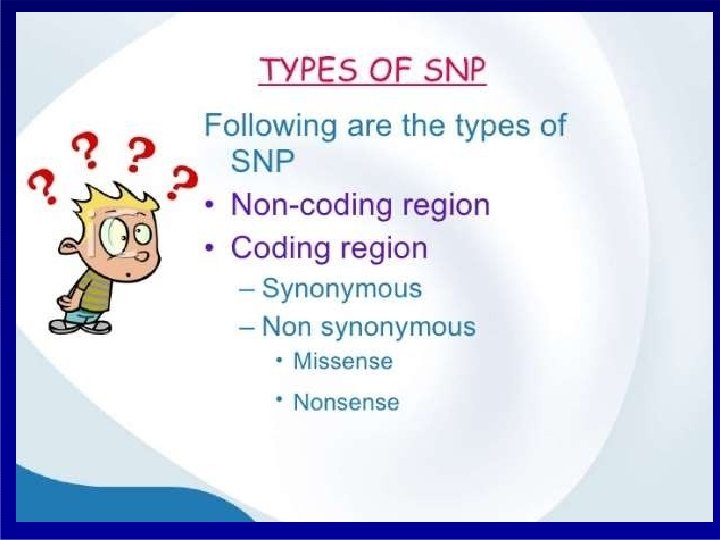
Techniques: Non-PCR Based, RFLP- Restriction fragment length polymorphism. PCR Based RAPD- Random amplification of polymorphic DNA. AFLP-Amplified fragment length polymorphism. SCAR-Sequence characterize amplified region. STS- Sequence tagged sites. EST-Express sequence tags. SNP-Single nucleotide polymorphism. SSR-Simple sequence repeats CAPS-Cleaved amplified polymorphic sequences.

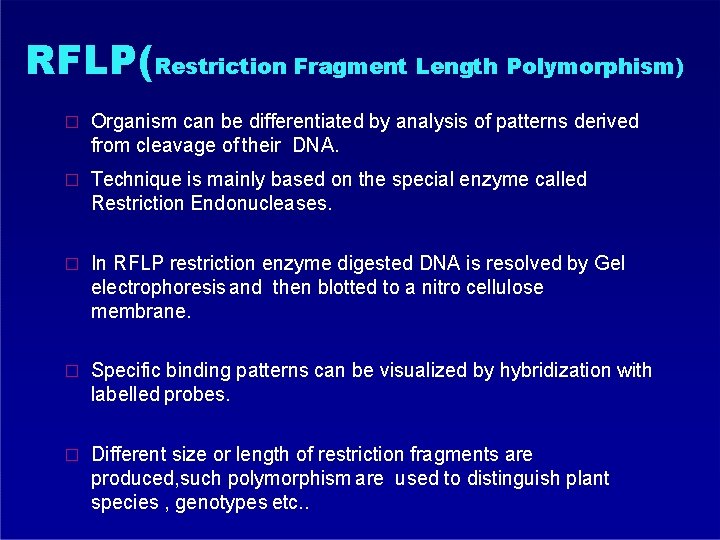
RFLP(Restriction Fragment Length Polymorphism) � Organism can be differentiated by analysis of patterns derived from cleavage of their DNA. � Technique is mainly based on the special enzyme called Restriction Endonucleases. � In RFLP restriction enzyme digested DNA is resolved by Gel electrophoresis and then blotted to a nitro cellulose membrane. � Specific binding patterns can be visualized by hybridization with labelled probes. � Different size or length of restriction fragments are produced, such polymorphism are used to distinguish plant species , genotypes etc. .

Application � Used in phylogenetic studies. � Gene mapping. � DNA finger printing. � Studies of gene flow

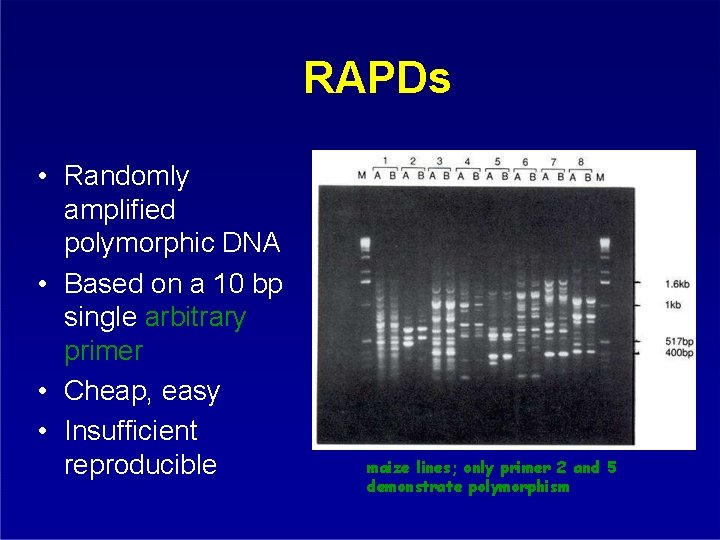
RAPDs • Randomly amplified polymorphic DNA • Based on a 10 bp single arbitrary primer • Cheap, easy • Insufficient reproducible maize lines; only primer 2 and 5 demonstrate polymorphism

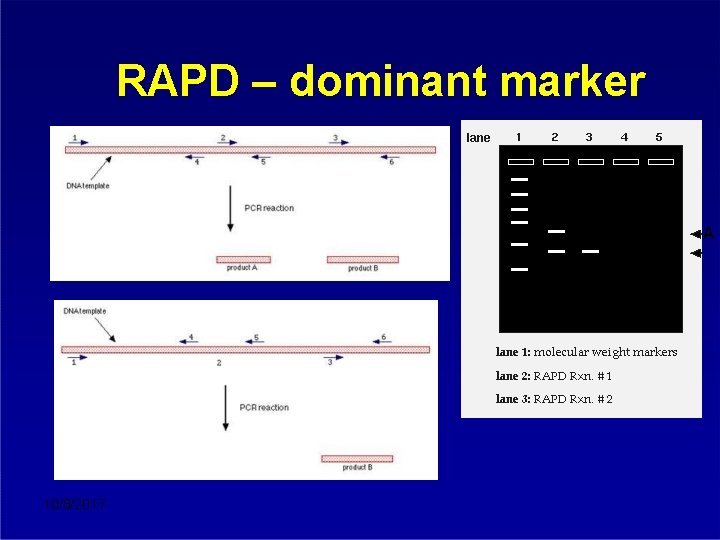
RAPD – dominant marker A 10/8/2017

AFLP(Amplified Fragment Length Polymorphism) � AFLP is based on a selectively amplifying a subset of restriction fragments from a complex mixture of DNA fragments obtained after digestion of genomic DNA with restriction endonucleases. � The technique involves four steps: (1) Restriction of DNA. (2) Ligation of oligonucleotide adapters. (3) Amplification. (4) Gel analysis of amplified fragments. � These fragments are viewed on denaturing polyacrylamide gels either through autoradiographic or fluorescence methodologies.

Application � Applied in studies involving genetic identity, identification of clones and cultivars. � phylogenetic studies of closely related species. � This technique is useful for breeders to accelerate plant improvement. � AFLP markers are useful in genetic studies, such as biodiversity evaluation, analysis of germplasm collections, genotyping of individuals and genetic distance analyses.

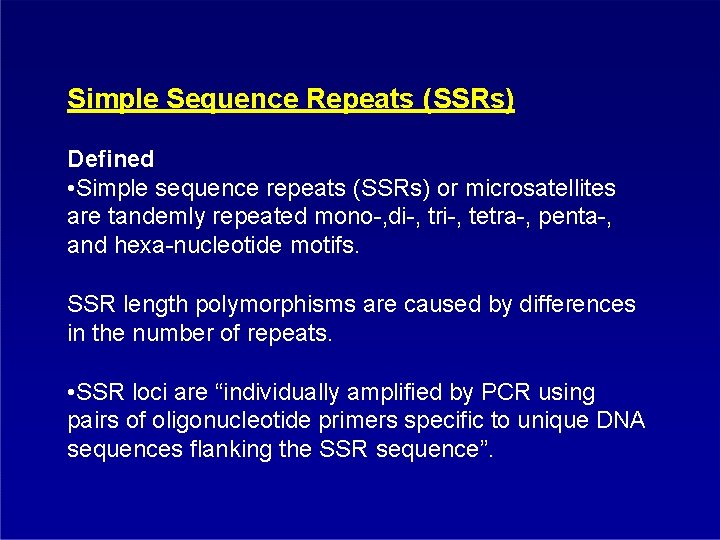
Simple Sequence Repeats (SSRs) Defined • Simple sequence repeats (SSRs) or microsatellites are tandemly repeated mono-, di-, tri-, tetra-, penta-, and hexa-nucleotide motifs. SSR length polymorphisms are caused by differences in the number of repeats. • SSR loci are “individually amplified by PCR using pairs of oligonucleotide primers specific to unique DNA sequences flanking the SSR sequence”.

Why Have SSRs Had Such a Large Impact on Genomics? • SSRs tend to be highly polymorphic. • SSRs are highly abundant and randomly dispersed throughout most genomes. • Most SSR markers are co-dominant and locus specific. • Genotyping throughput is high and can be automated.

SSR / microsatellite 1. Isolation of DNA fragments (vectors) containing a simple sequence repeat (microsatellite), e. g. [AT]n [GC]n, [CGA]n, [GATA]n 1. Sequencing regions flanking the SSR 2. Designing primers for border sequences 3. Testing in population for duplications and SSR polymorphism
![SSR methodolgy Genotype A Genotype B AT1 8 AT2 2 AT1 AT2 PCR SSR - methodolgy Genotype A Genotype B [AT]1 8 [AT]2 2 [AT]1 [AT]2 PCR](https://slidetodoc.com/presentation_image_h2/41221deccec3d7ab92acdb34433742d0/image-24.jpg)
SSR - methodolgy Genotype A Genotype B [AT]1 8 [AT]2 2 [AT]1 [AT]2 PCR amplification with 2 8 radiolabelled nucleoltide Polyacrylamide Gel Electrophoresis of PCR products and autoradiography Genotype A Genotype B

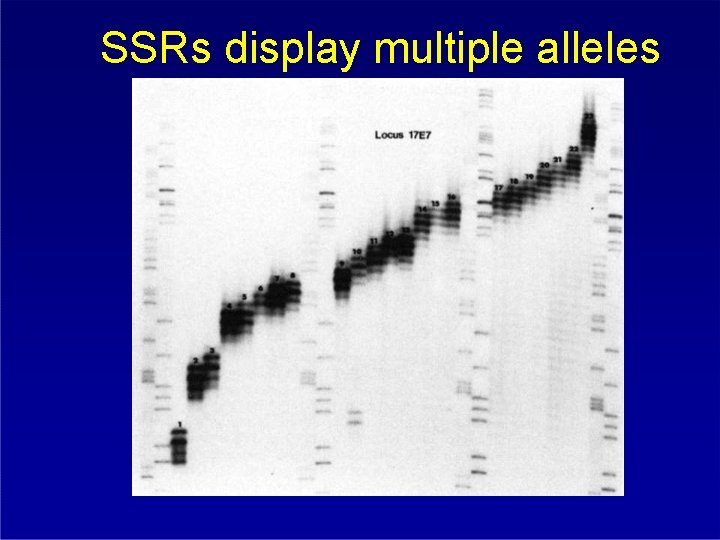
SSRs display multiple alleles

10/8/2017 NIBGE Ph. D lecture

10/8/2017 NIBGE Ph. D lecture

10/8/2017 NIBGE Ph. D lecture

10/8/2017 NIBGE Ph. D lecture

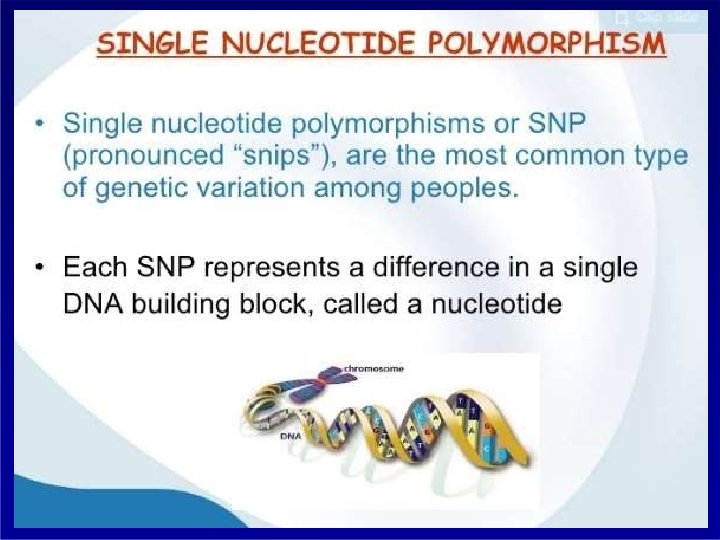
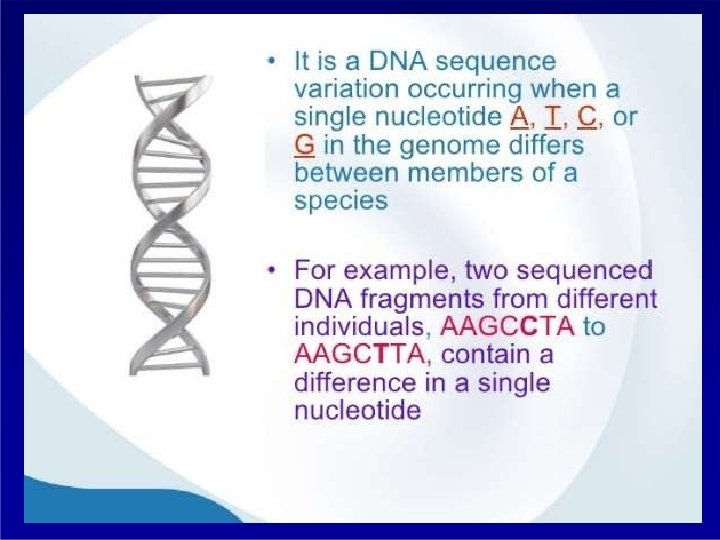
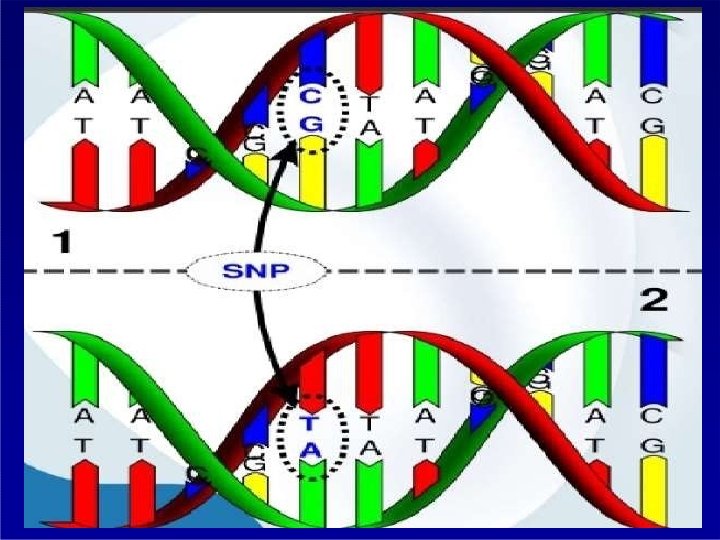
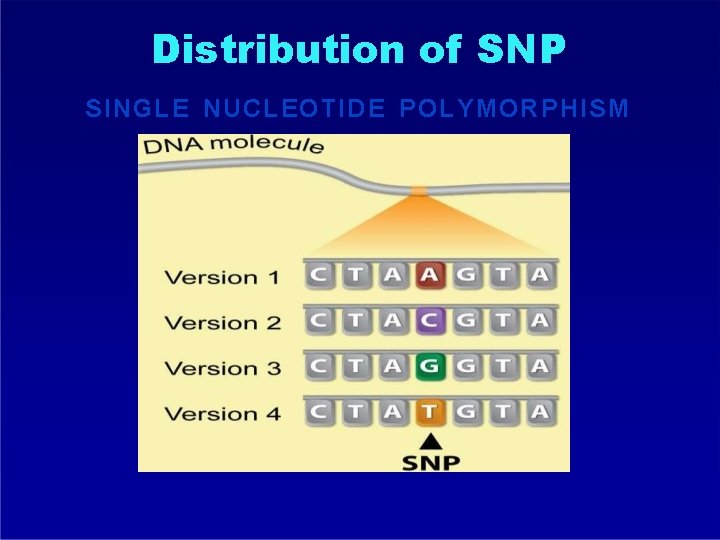
Distribution of SNP SINGLE NUCLEOTIDE POLYMORPHISM

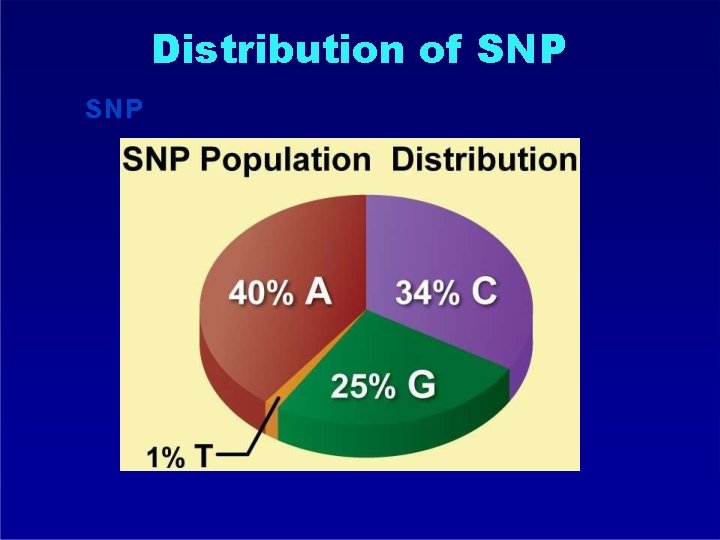
Distribution of SNP

10/8/2017 NIBGE Ph. D lecture

10/8/2017 NIBGE Ph. D lecture

10/8/2017 NIBGE Ph. D lecture

10/8/2017 NIBGE Ph. D lecture

10/8/2017 NIBGE Ph. D lecture

10/8/2017 NIBGE Ph. D lecture

10/8/2017 NIBGE Ph. D lecture
 Hot site cold site warm site disaster recovery
Hot site cold site warm site disaster recovery There is there are
There is there are Any to any connectivity
Any to any connectivity Seknder
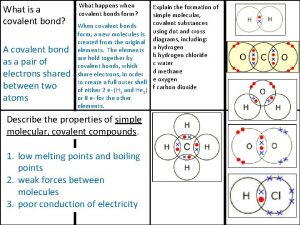
Seknder Covalent bond boiling point
Covalent bond boiling point Ionic covalent metallic
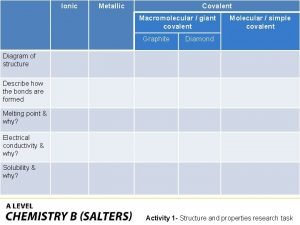
Ionic covalent metallic Giant molecular structure vs simple molecular structure
Giant molecular structure vs simple molecular structure Hepatitis comparison
Hepatitis comparison Thin layers of volcanic ash act as excellent time markers
Thin layers of volcanic ash act as excellent time markers Taboo words
Taboo words Mrs midas 10 marker
Mrs midas 10 marker Floating red markers are called nuns. what shape are nuns
Floating red markers are called nuns. what shape are nuns Ethnic boundary marker
Ethnic boundary marker What is surrogate marker
What is surrogate marker Stansw meet the markers
Stansw meet the markers Discourse markers examples
Discourse markers examples Greed
Greed Ca 125
Ca 125 White diamond marking on the roadway indicates
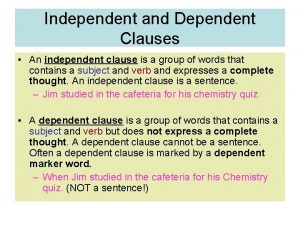
White diamond marking on the roadway indicates Independent clause
Independent clause Gramene marker database
Gramene marker database Rflp vs rapd
Rflp vs rapd Gcse pe 9 marker
Gcse pe 9 marker Asl discourse markers
Asl discourse markers Example of discourse markers
Example of discourse markers Discourse markers
Discourse markers St andrews eap conference
St andrews eap conference Volcanic ash
Volcanic ash 1000 foot markers runway
1000 foot markers runway Written discourse markers
Written discourse markers Restrictive adjective clause examples
Restrictive adjective clause examples Stance markers
Stance markers What is biological marker
What is biological marker Discourse markers result
Discourse markers result Lie past perfect tense
Lie past perfect tense Dr. penina segall-gutierrez
Dr. penina segall-gutierrez What are voice markers they say i say
What are voice markers they say i say When to use past perfect continuous
When to use past perfect continuous No passing zone
No passing zone