Mapping of Simple Complex Genetic Diseases Anne Haake











- Slides: 28

Mapping of Simple & Complex Genetic Diseases Anne Haake Rhys Price Jones

Simple Diseases • Follow Mendelian inheritance patterns – e. g. autosomal dominant, x-linked recessive • Generally rare • Caused by changes in one gene • Examples: Cystic Fibrosis, Duchenne Muscular Dystrophy

Complex Diseases • aka Common Diseases • Tend to cluster in families but do not follow Mendelian inheritance patterns • Result from action of multiple genes • Alleles of these genes are “susceptibility factors” • Most factors are neither necessary or sufficient for disease • Complex interaction between environment and these susceptibility alleles contributes to disease

Complex Diseases • Examples: diabetes, asthma, cardiovascular disease, many cancers, high blood pressure, Alzheimer’s disease • Many more. .

How do we study these? • Simple diseases: – Usually a complete correlation between genotype and phenotype – “easy” to analyze • A nice overview of strategies by Dennis Drayna at NHGRI • http: //www. nhgri. nih. gov/Pages/Hyperion/CO URSE 2000/Pdf/Drayna. pdf

Positional Cloning Approach • Isolate a disease gene based on its chromosomal position • No prior knowledge of structure, function, or pathological mechanism

Need some markers • DNA polymorphisms “many forms” • Variation in population allows us to use them as informative markers • Identified by common lab techniques such as PCR • Examples: – RFLP-restriction length polymorphisms – Microsatellites- tandem repeats, e. g (CA)n – SNPs-single nucleotide polymorphisms

Recombination Frequency • RF (genetic distance), also called q (theta) between 2 loci is related to how far apart they are on the chromosome (physical distance) • So. . can estimate physical distances by measuring q. • 1% RF roughly equivalent to 1 c. M (1 Mb DNA) http: //www. abdn. ac. uk/~gen 155/lectures/gn 380 1 b. htm#ls

Strategy • Look for co-inheritance of disease and some marker; known as linkage • If a marker (polymorphism) is close to a disease gene then there is a low chance of meiotic recombination between them • Family studies are required; study of individuals in generations allows us to figure out pattern of inheritance of disease relative to markers • Generate LOD Scores http: //www. ndsu. nodak. edu/instruct/mcclean/plsc 431/lin kage/linkage 6. htm

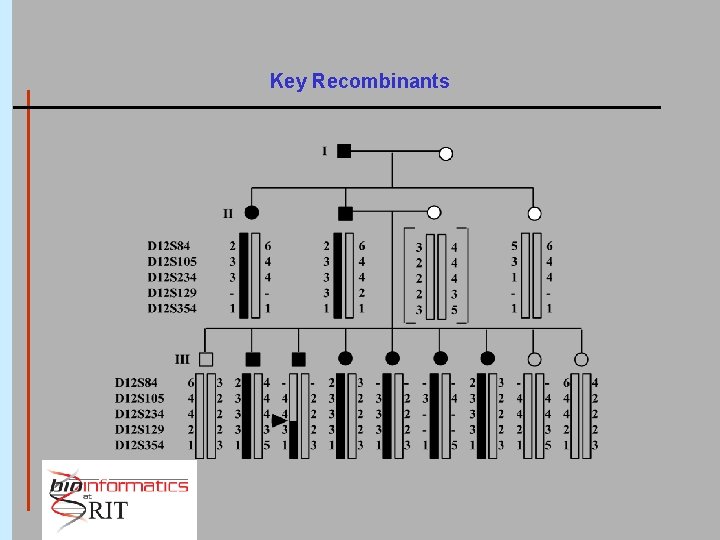
An Example: Darier's Disease Synonyms: Mc. Kusick #12420 Darier-White Disease Keratosis follicularis Genetics: autosomal dominant high penetrance 1: 100, 000 Denmark 1: 36, 000 northeast England

Key Recombinants

Genes Mapped to 12 q 23 -24. 1 IGFInsulin-like Growth Factor NFYB Nuclear Factor Binding to Y PAH Phenylalanine Hydroxlyase TSC 3 Tuberous Sclerosis ACADS acyl-coenzyme A dehydrogenase ATP 2 A 2 ATPase Ca++ transporting SCA 2 Spinal Cerebellar Ataxia MYL 2 Myosin light polypeptide PMCH pro-melanin-concentration PLA 2 A Phospholipase 2 A IFNG Interferon gamma PPP 1 CC Protein phosphatase 1 ALDH 2 Aldehyde dehydrogenase NOS 1 Nitric Oxide Synthase TRA 1 Tumor Rejection Antigen ZNF 26 Zinc Finger Protein TCF 1 Transcription Factor 1 UBC Ubiquitin C SPSMA Scapuloperoneal spinal muscular atrophy

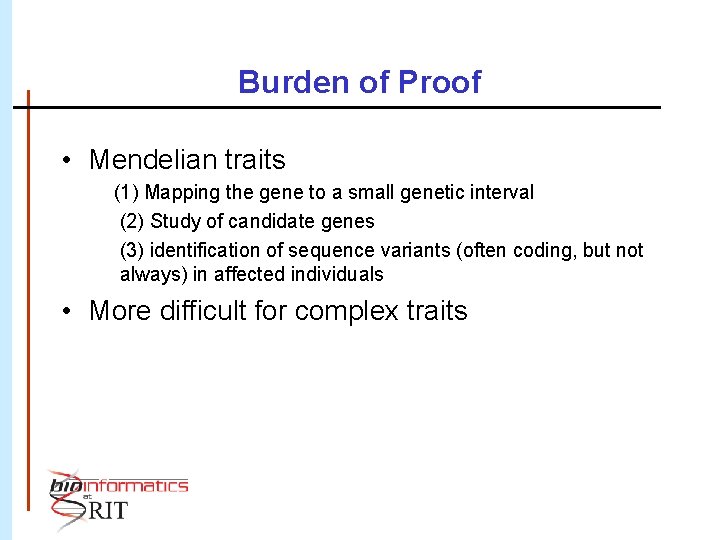
Burden of Proof • Mendelian traits (1) Mapping the gene to a small genetic interval (2) Study of candidate genes (3) identification of sequence variants (often coding, but not always) in affected individuals • More difficult for complex traits

Quantitative Trait Loci (QTL) • Complex traits are also known as QTLs • Term used most in agricultural, horticultural genetics • Why quantitative? • Consider Mendelian traits – Cross short pea plant vs. tall pea plant – F 2 generation: you know the genotype of the short plants and you can generalize the genotype of the tall & can predict phenotype from genotype – Phenotypes are called discontinuous traits

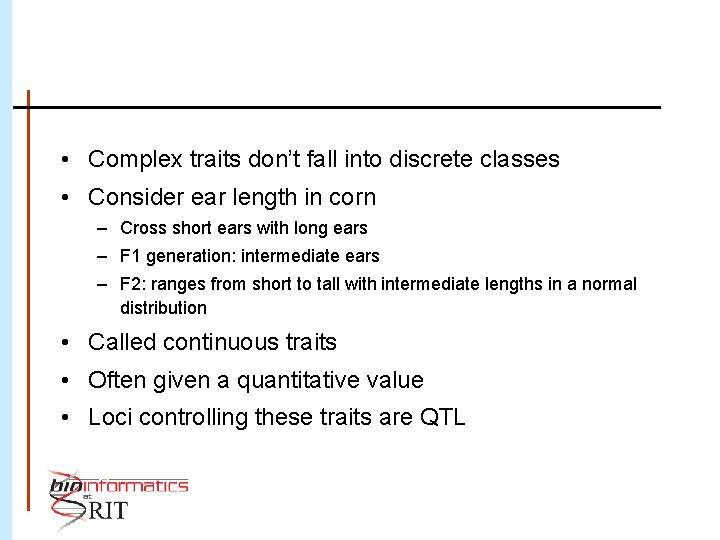
• Complex traits don’t fall into discrete classes • Consider ear length in corn – Cross short ears with long ears – F 1 generation: intermediate ears – F 2: ranges from short to tall with intermediate lengths in a normal distribution • Called continuous traits • Often given a quantitative value • Loci controlling these traits are QTL

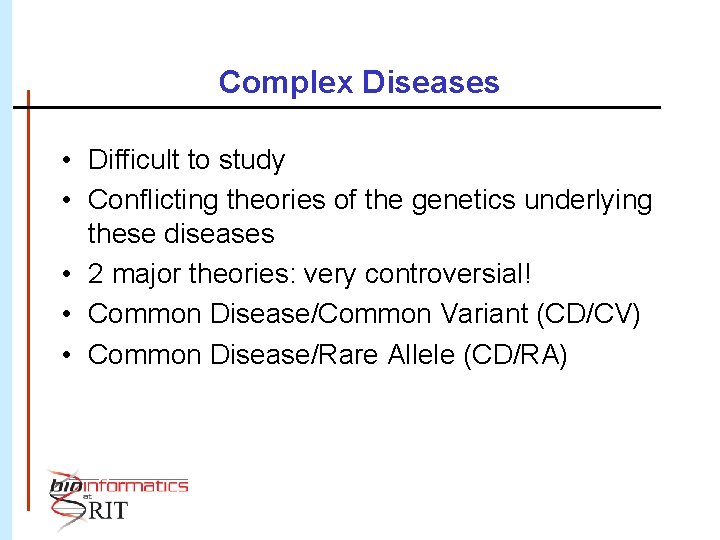
Complex Diseases • Difficult to study • Conflicting theories of the genetics underlying these diseases • 2 major theories: very controversial! • Common Disease/Common Variant (CD/CV) • Common Disease/Rare Allele (CD/RA)

• CD/CV – Alleles that existed prior to the global dispersal of humans or those subject to positive selection represent a significant proportion of the susceptibility alleles for common disease • CD/RA – Most mutations underlying common disease have occurred after the divergence of populations – Expect heterogeneity in genes in common diseases

CD/CV • Susceptibility alleles confer moderate risk and occur at relatively high rates in the population (>= 1%). • Suggests that association studies in large cohort populations (e. g. unrelated individuals sharing the common disease) will be fruitful • SNPs have facilitated this type of study – easy to measure, stable in population

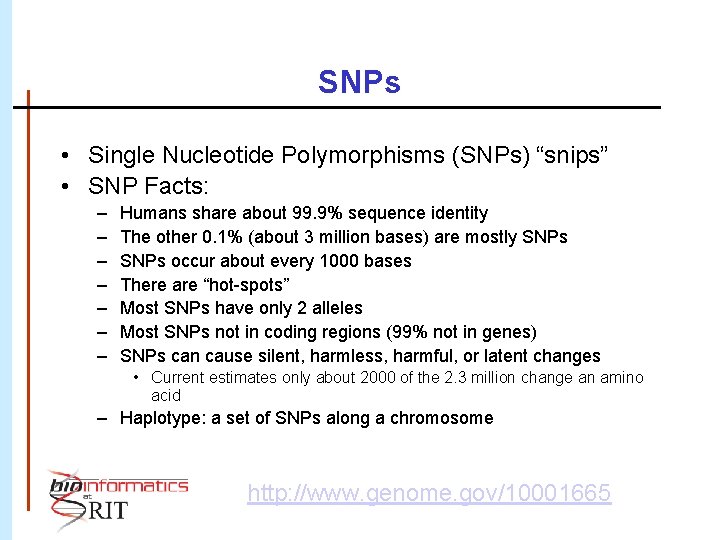
SNPs • Single Nucleotide Polymorphisms (SNPs) “snips” • SNP Facts: – – – – Humans share about 99. 9% sequence identity The other 0. 1% (about 3 million bases) are mostly SNPs occur about every 1000 bases There are “hot-spots” Most SNPs have only 2 alleles Most SNPs not in coding regions (99% not in genes) SNPs can cause silent, harmless, harmful, or latent changes • Current estimates only about 2000 of the 2. 3 million change an amino acid – Haplotype: a set of SNPs along a chromosome http: //www. genome. gov/10001665

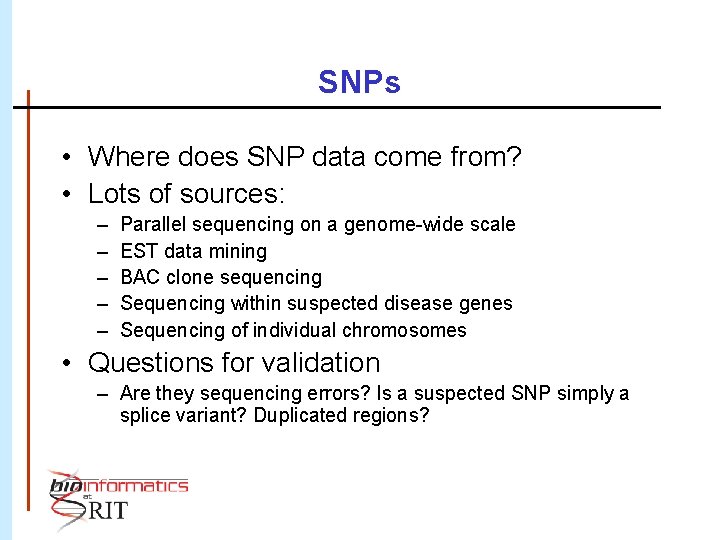
SNPs • Where does SNP data come from? • Lots of sources: – – – Parallel sequencing on a genome-wide scale EST data mining BAC clone sequencing Sequencing within suspected disease genes Sequencing of individual chromosomes • Questions for validation – Are they sequencing errors? Is a suspected SNP simply a splice variant? Duplicated regions?

Association Studies • SNPs usually serve as biological markers rather than underlying cause of disease • SNP is located near a gene associated with a disease • Allelic association aka linkage disequilibrium • Compare genome wide SNP profiles from individuals with the disease to those without the disease. • Difference identifies a putative disease profile that may eventually be used in diagnosis

Haplotype Mapping • Definition of a complete Hap. Map one of the goals of the SNP Consortium • Questions remain in the community about the degree of linkage disequilibrium in the human population • Estimates vary from 3 kb-400 kb • Not very useful for disease mapping at either end

Burden of Proof • Complex Diseases-what are the steps to gene discovery? (1) Linkage or Association -challenges in testing numerous genetic markers for linkage and correlating inheritance patterns -minimal intervals of QTLs are usually no less than 10 -30 c. M (typically 100 -300 genes in that interval) -makes candidate gene studies difficult

Burden of Proof for Complex Diseases • (2) Fine-mapping – Genetic crosses, family-based studies of linkage disequilibrium using dense markers – Are SNPs the optimal markers? • (3) Sequence analysis to identify candidate variants • (4) Functional tests such as replacement of variant to swap phenotypes • (5) Additional evidence at cellular and tissue levels

Model Organisms • One of most promising approaches is to extend the human mapping studies to animal models • Take advantage of highly inbred strains • Take advantage of genome synteny to relate mouse results back to human genes.

Successful Use of Genome-Wide Screens • Alzheimer’s disease – Apo. E gene has 2 SNPs – 3 alleles Apo. E 2, Apo. E 3, Apo. E 4 – Association of the Apo. E 4 allele with Alzheimer’s disease & APOE 4 protein in brain lesions • Mouse: mutations in tubby gene – Cause obesity, retinal degeneration, hearing loss – More evidence of multi-gene interactions • Modifier gene (moth 1) protects tubby mice from hearing loss • Mtap 1 a c. DNA rescues hearing loss

High-throughput SNP analysis • Genotyping via oligonucleotide arrays • e. g. Affymetrix has 10 K and 100 K arrays • Analysis with DNA isolated from only a few drops of blood

Data Analysis? • Shares some problems with gene expression arrays – e. g. get measurements across many, many genes • Some use of clustering/classification approaches to discover patterns in the data