Human Genetics Concepts and Applications Tenth Edition RICKI















- Slides: 40

Human Genetics Concepts and Applications Tenth Edition RICKI LEWIS 14 Constant Allele Frequencies Power. Point® Lecture Outlines Prepared by Johnny El-Rady, University of South Florida Copyright ©The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display

Introduction Population = An interbreeding group of the same species in a given geographical area Gene pool = The collection of alleles in the members of the population Population genetics = The study of the genetics of a population and how the alleles vary with time Gene Flow = Movement of alleles between populations when people migrate and mate 2

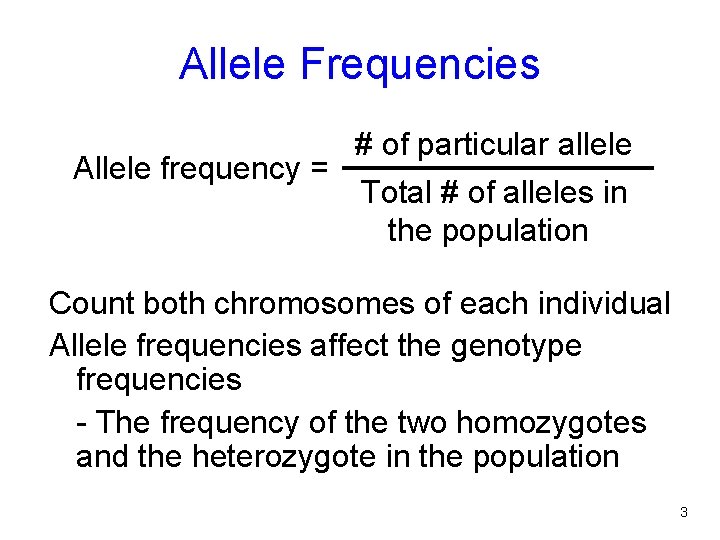
Allele Frequencies Allele frequency = # of particular allele Total # of alleles in the population Count both chromosomes of each individual Allele frequencies affect the genotype frequencies - The frequency of the two homozygotes and the heterozygote in the population 3

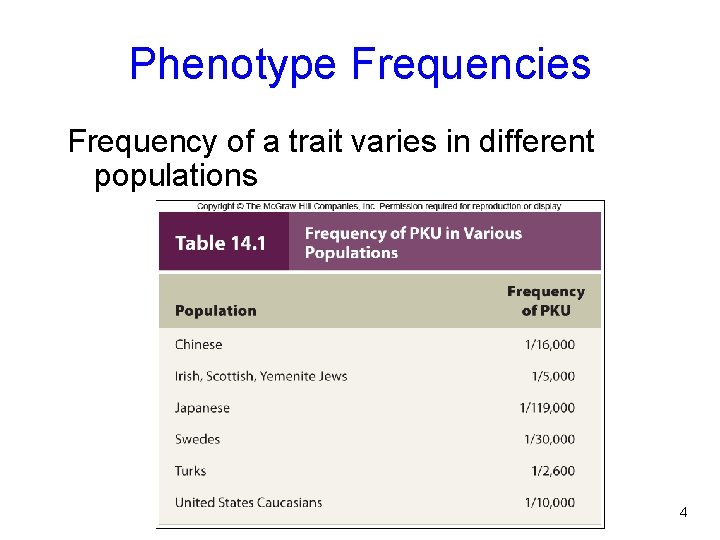
Phenotype Frequencies Frequency of a trait varies in different populations Table 14. 1 4

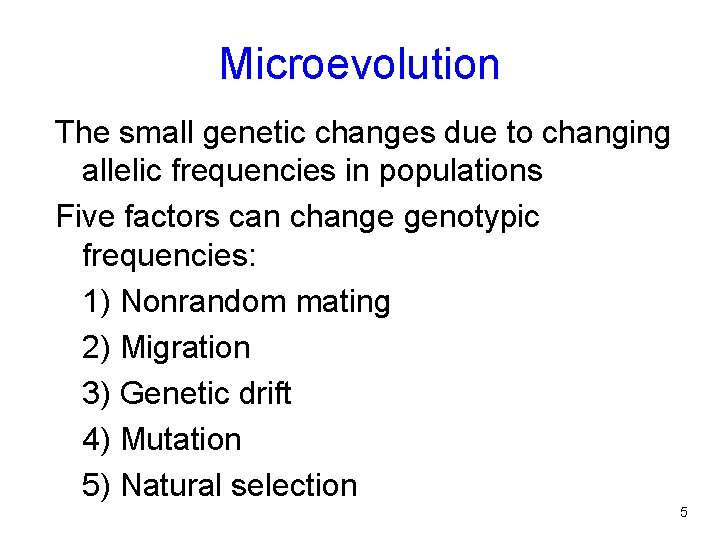
Microevolution The small genetic changes due to changing allelic frequencies in populations Five factors can change genotypic frequencies: 1) Nonrandom mating 2) Migration 3) Genetic drift 4) Mutation 5) Natural selection 5

Macroevolution Refers to the formation of new species Occurs when enough microevolutionary changes have occurred to prevent individuals from one population to successfully produce fertile offspring 6

Hardy-Weinberg Equation Developed independently by an English mathematician and a German physician Used algebra to explain how allele frequencies predict genotypic and phenotypic frequencies in a population of diploid, sexually-reproducing species Disproved the assumption that dominant traits would become more common, while recessive traits would become rarer 7

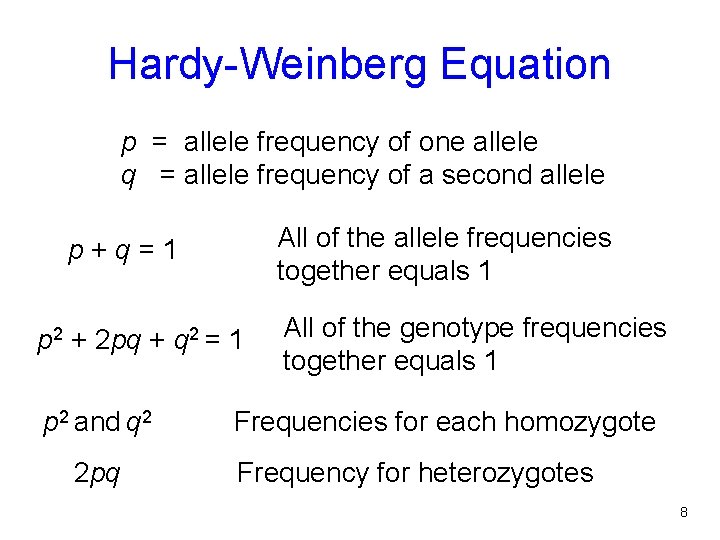
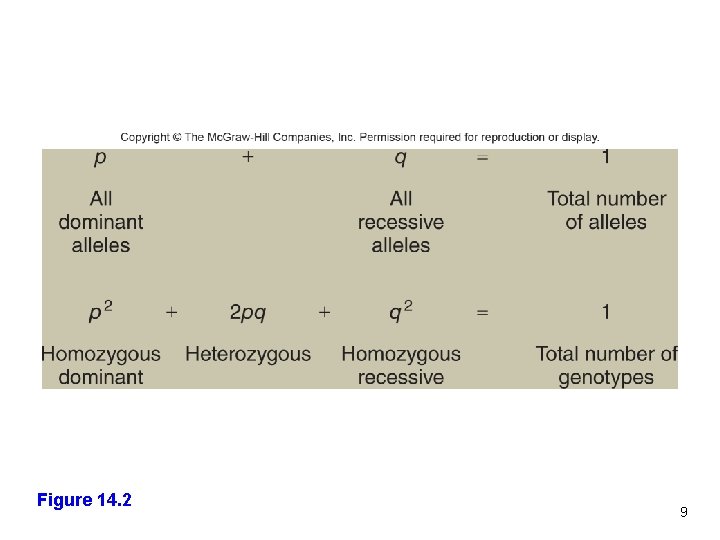
Hardy-Weinberg Equation p = allele frequency of one allele q = allele frequency of a second allele All of the allele frequencies together equals 1 p+q=1 p 2 + 2 pq + q 2 = 1 p 2 and q 2 2 pq All of the genotype frequencies together equals 1 Frequencies for each homozygote Frequency for heterozygotes 8

Figure 14. 2 9

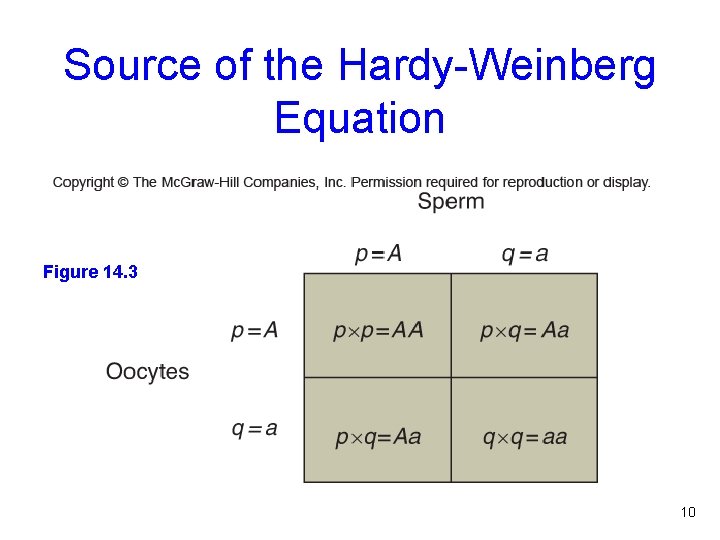
Source of the Hardy-Weinberg Equation Figure 14. 3 10

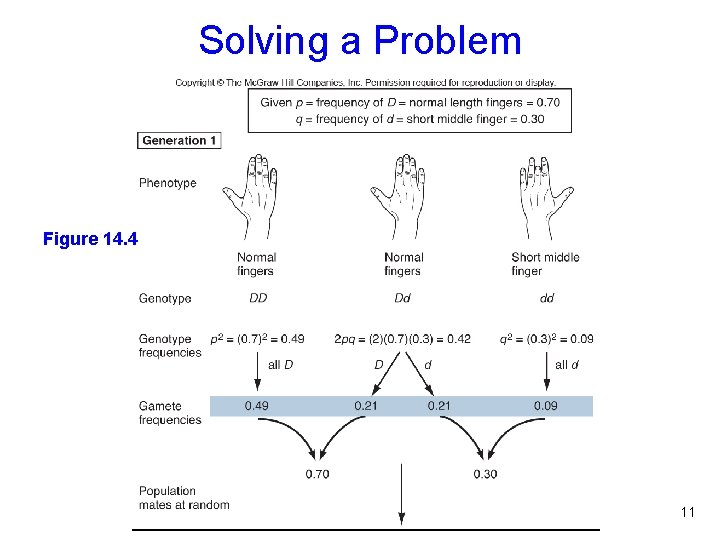
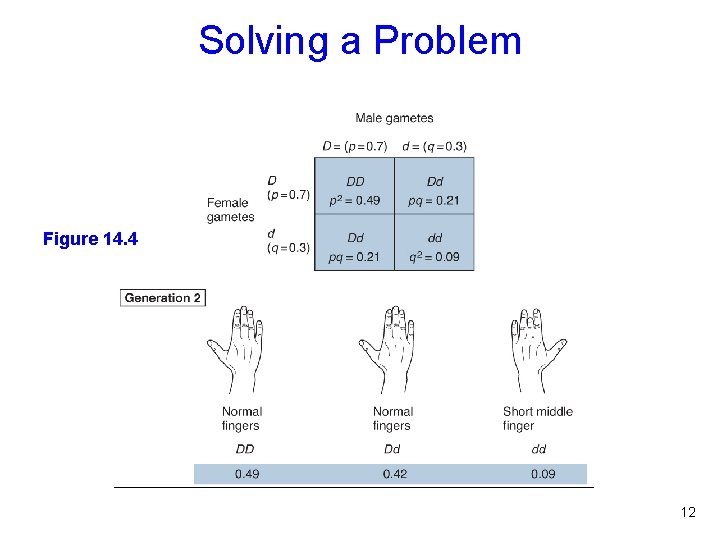
Solving a Problem Figure 14. 4 11

Solving a Problem Figure 14. 4 12

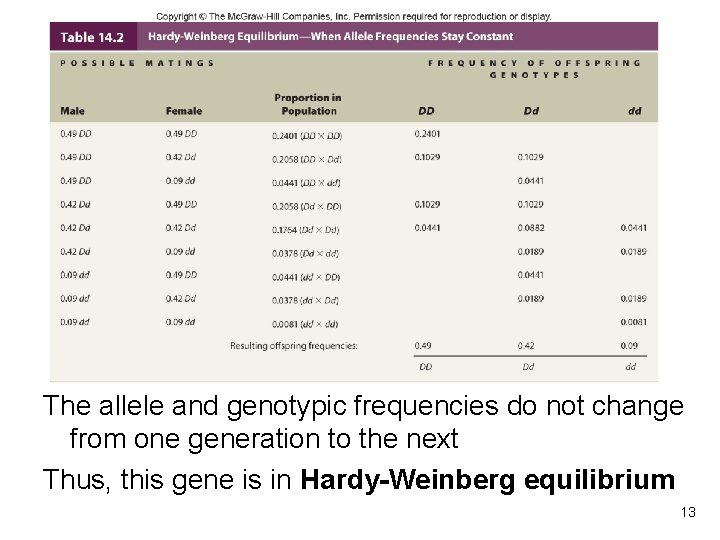
The allele and genotypic frequencies do not change from one generation to the next Thus, this gene is in Hardy-Weinberg equilibrium Table 14. 2 13

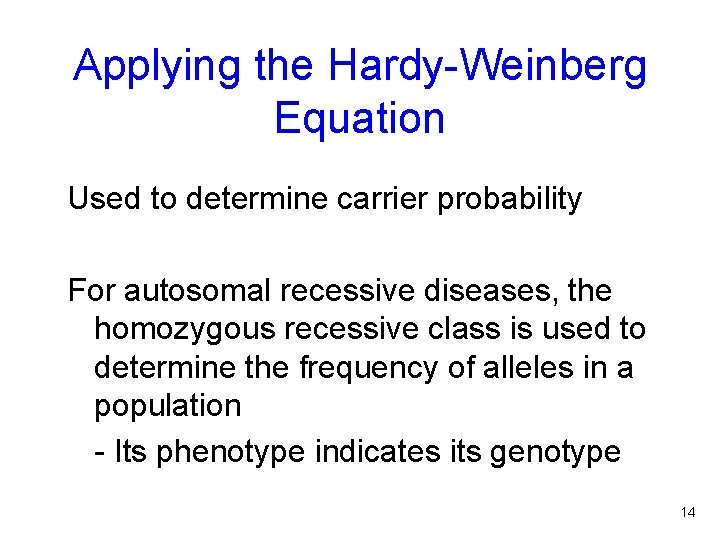
Applying the Hardy-Weinberg Equation Used to determine carrier probability For autosomal recessive diseases, the homozygous recessive class is used to determine the frequency of alleles in a population - Its phenotype indicates its genotype Figure 14. 3 14

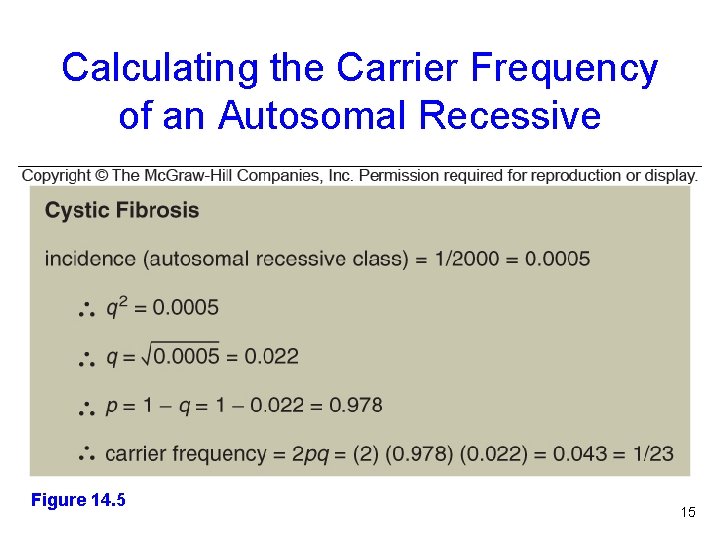
Calculating the Carrier Frequency of an Autosomal Recessive Figure 14. 5 Figure 14. 3 15

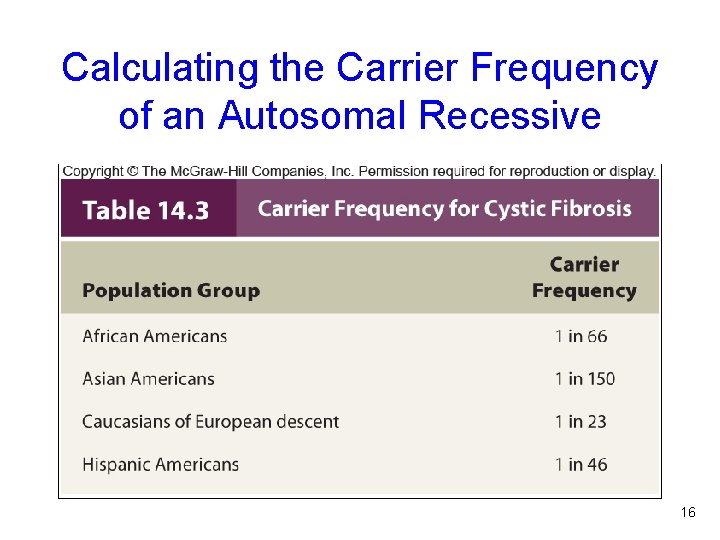
Calculating the Carrier Frequency of an Autosomal Recessive Table 14. 3 16

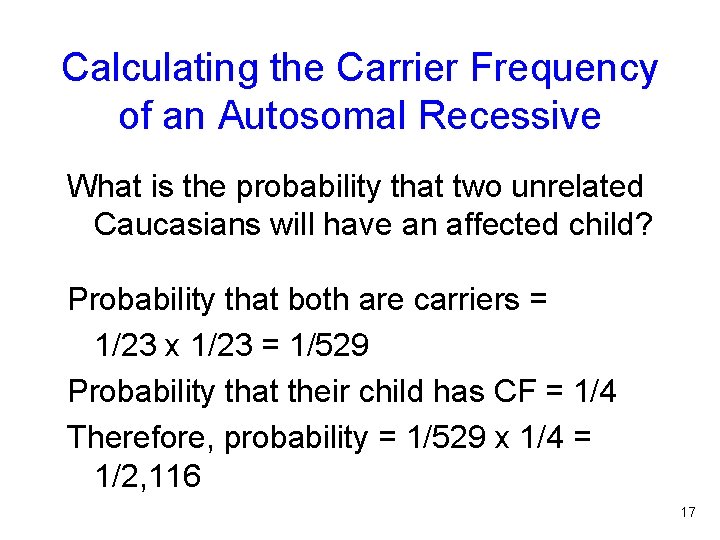
Calculating the Carrier Frequency of an Autosomal Recessive What is the probability that two unrelated Caucasians will have an affected child? Probability that both are carriers = 1/23 x 1/23 = 1/529 Probability that their child has CF = 1/4 Therefore, probability = 1/529 x 1/4 = 1/2, 116 Figure 14. 3 17

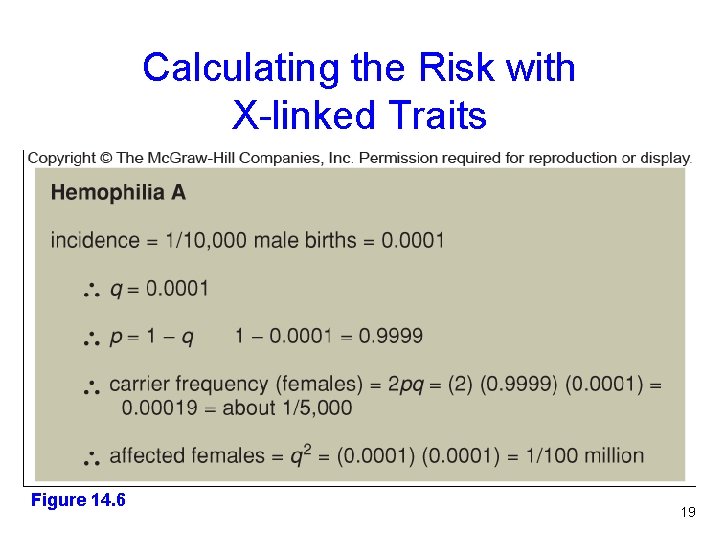
Calculating the Risk with X-linked Traits For females, the standard Hardy-Weinberg equation applies p 2 + 2 pq + q 2 = 1 However, in males the allele frequency is the phenotypic frequency p + q= 1 Figure 14. 3 18

Calculating the Risk with X-linked Traits Figure 14. 6 19

Hardy-Weinberg Equilibrium Hardy-Weinberg equilibrium is rare for protein-encoding genes that affect the phenotype However, it does apply to portions of the genome that do not affect phenotype These include repeated DNA segments - Not subject to natural selection 20

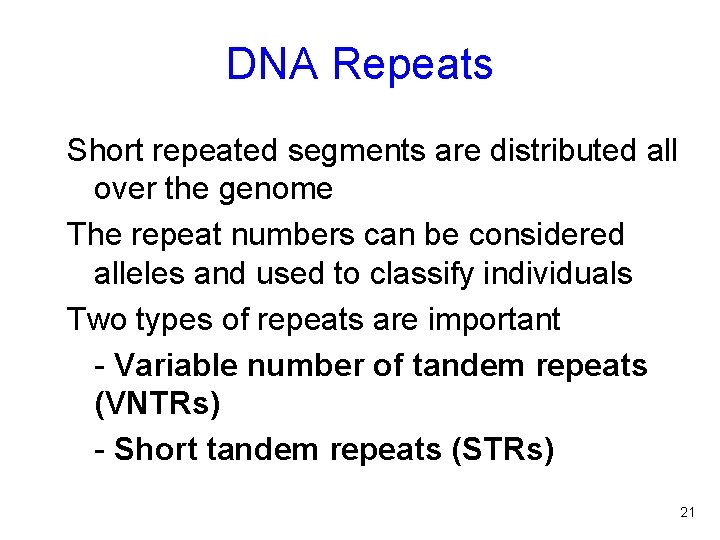
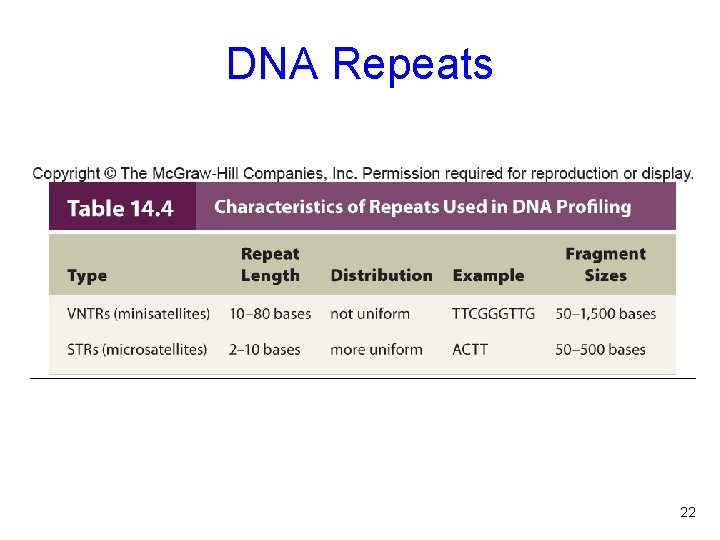
DNA Repeats Short repeated segments are distributed all over the genome The repeat numbers can be considered alleles and used to classify individuals Two types of repeats are important - Variable number of tandem repeats (VNTRs) - Short tandem repeats (STRs) 21

DNA Repeats 22

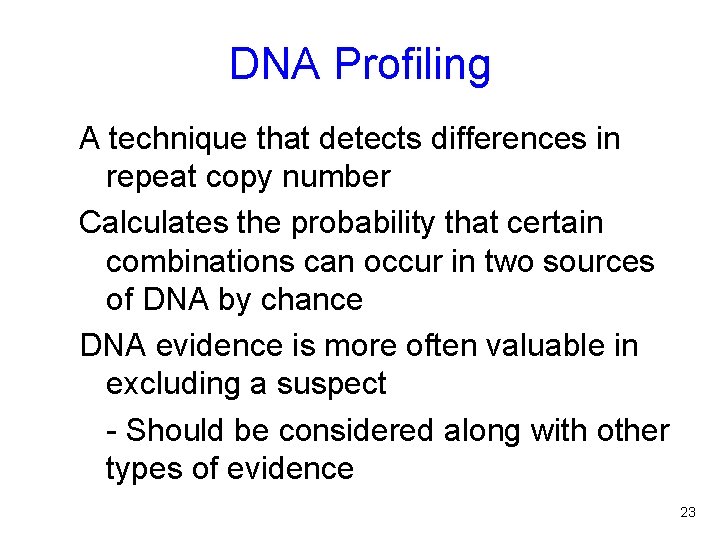
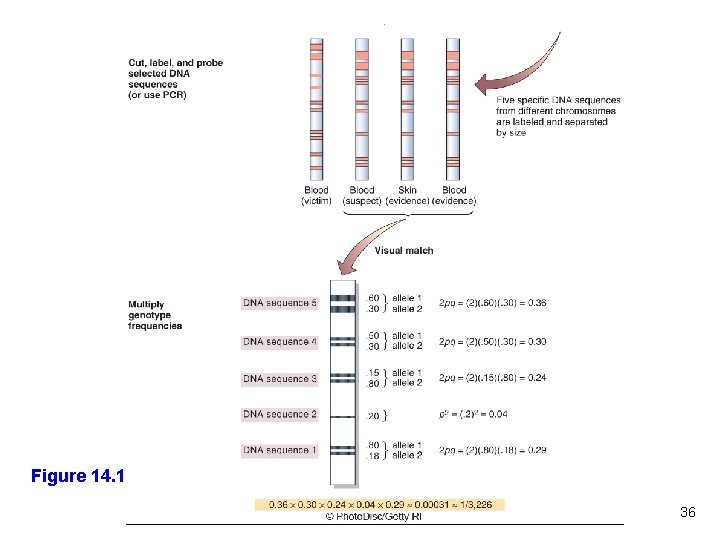
DNA Profiling A technique that detects differences in repeat copy number Calculates the probability that certain combinations can occur in two sources of DNA by chance DNA evidence is more often valuable in excluding a suspect - Should be considered along with other types of evidence 23

Comparing DNA Repeats Figure 14. 7 24

DNA Profiling Developed in the 1980 s by British geneticist Sir Alec Jeffreys Also called DNA fingerprinting Identifies individuals Used in forensics, agriculture, paternity testing, and historical investigations 25

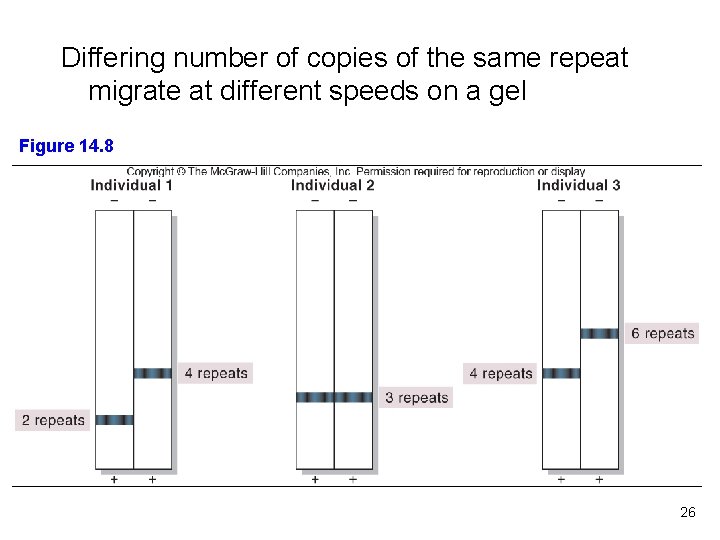
Differing number of copies of the same repeat migrate at different speeds on a gel Figure 14. 8 26

Jeffreys used his technique to demonstrate that Dolly was truly a clone of the 6 year old ewe that donated her nucleus Figure 14. 9 27

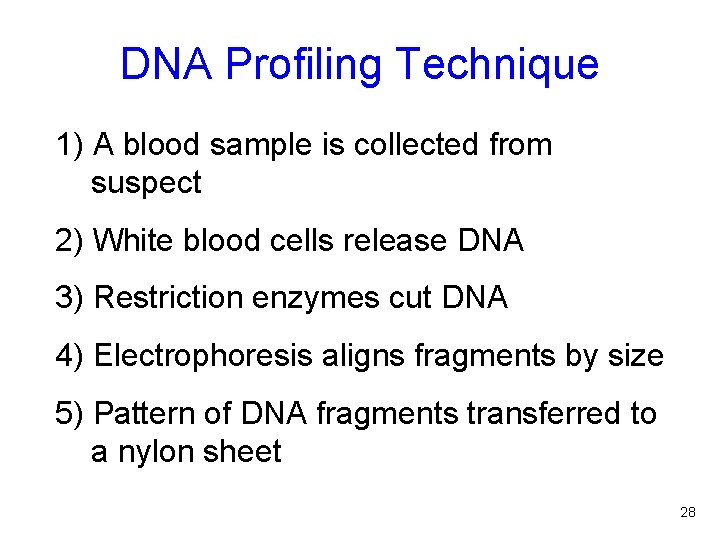
DNA Profiling Technique 1) A blood sample is collected from suspect 2) White blood cells release DNA 3) Restriction enzymes cut DNA 4) Electrophoresis aligns fragments by size 5) Pattern of DNA fragments transferred to a nylon sheet 28

DNA Profiling Technique 6) Exposed to radioactive probes 7) Probes bind to DNA 8) Sheet placed against X ray film 9) Pattern of bands constitutes DNA profile 10) Identify individuals 29

Box Figure 14. 1 30

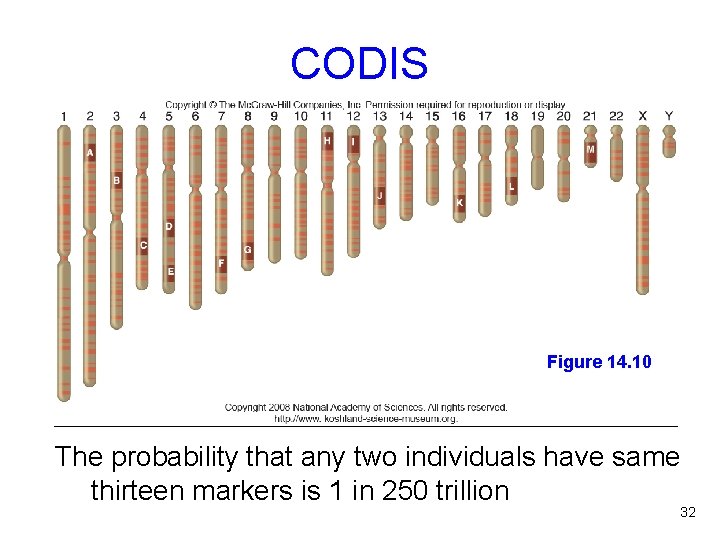
DNA Sources DNA can be obtained from any cell with a nucleus STRs are used when DNA is scarce If DNA is extremely damaged, mitochondrial DNA (mt. DNA) is often used For forensics, the FBI developed the Combined DNA Index System (CODIS) - Uses 13 STRs 31

CODIS Figure 14. 10 The probability that any two individuals have same thirteen markers is 1 in 250 trillion 32

Population Statistics Is Used to Interpret DNA Profiles The power of DNA profiling is greatly expanded by tracking repeats in different chromosomes The number of copies of a repeat are assigned probabilities based on their observed frequency in a population The product rule is then used to calculate probability of a certain repeat combination 33

To Catch A Thief With A Sneeze Table 14. 6 34

To Solve A Crime Table 14. 6 Figure 14. 11 35

Table 14. 6 Figure 14. 11 36

Using DNA Profiling to Identify Victims Recent examples of large-scale disasters - World Trade Center attack (2001) - Indian Ocean Tsunami (2004) - Hurricane Katrina (2005) 37

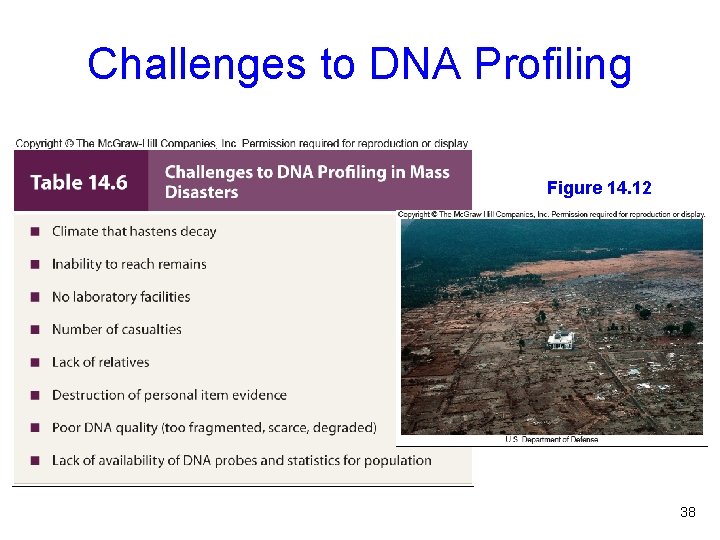
Challenges to DNA Profiling Figure 14. 12 38

Figure 14. 12 39

Genetic Privacy Today’s population genetics presents a powerful way to identify individuals Our genomes can vary in more ways than there are people in the world DNA profiling introduces privacy issues - Example: DNA dragnets 40