Genespecific Methylation Analyses Fade Gong Nam Nguyen Introduction


- Slides: 22

Gene-specific Methylation Analyses Fade Gong & Nam Nguyen

Introduction DNA methylation in mammals occurs at Cp. G sites, where it serves many functions: • Self vs. non-self identification (such as in viral/bacterial infection) • Target for further regulatory events; examples: • Maintenance methyltransferases (functions after replication on hemi-methylated DNA) • Proteins involved in chromatin folding • Cp. G islands are elements of most promoters, which typically downregulate transcription in the methylated state

Principles of DNA Methylation Analyses 1. Single-gene methylation analyses Sensitivity of restriction enzymes for methylated Cp. G sites b. Bisulfite-mediated conversion of DNA a. 2. Genome-wide methylation analyses a. Anti-methylcytidine Ab b. HPLC (high-performance liquid chromatography)/mass spectrometry

Overview: Single-gene methylation analyses a) Methylation-sensitive restriction enzymes • Southern-blot hybridization b) Bisulfite conversion of DNA • • • Sequencing Methylation-specific PCR (MSP) Real-time MSP COBRA Pyrosequencing Mass. ARRAY

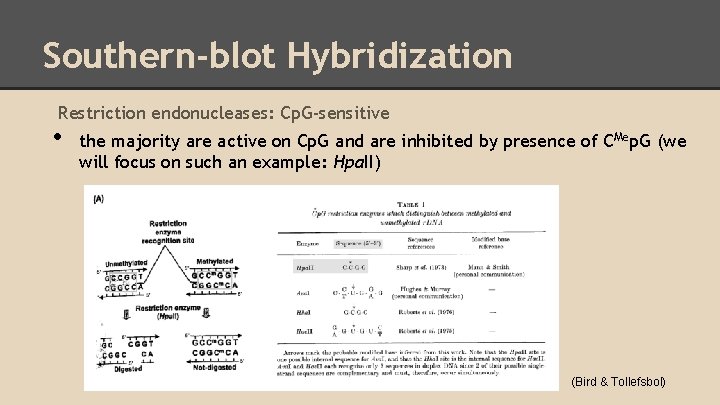
Southern-blot Hybridization Restriction endonucleases: Cp. G-sensitive • the majority are active on Cp. G and are inhibited by presence of CMep. G (we will focus on such an example: Hpa. II) (Bird & Tollefsbol)

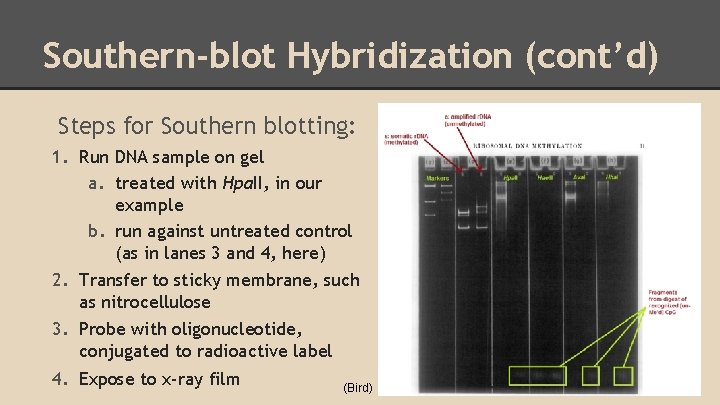
Southern-blot Hybridization (cont’d) Steps for Southern blotting: 1. Run DNA sample on gel a. treated with Hpa. II, in our example b. run against untreated control (as in lanes 3 and 4, here) 2. Transfer to sticky membrane, such as nitrocellulose 3. Probe with oligonucleotide, conjugated to radioactive label 4. Expose to x-ray film (Bird)

Southern-blot Hybridization (cont’d) Advantages 1. Relatively quantitative 2. Low cost Disadvantages 1. Low flexibility in selection of DNA region (i. e. must be isolated via other restriction enzymes that don’t degrade the fragment of interest) 2. Large amount of high-quality DNA required 3. Not all Cp. G’s presented are subjected to the assay (depends on selectivity of the restriction enzymes)

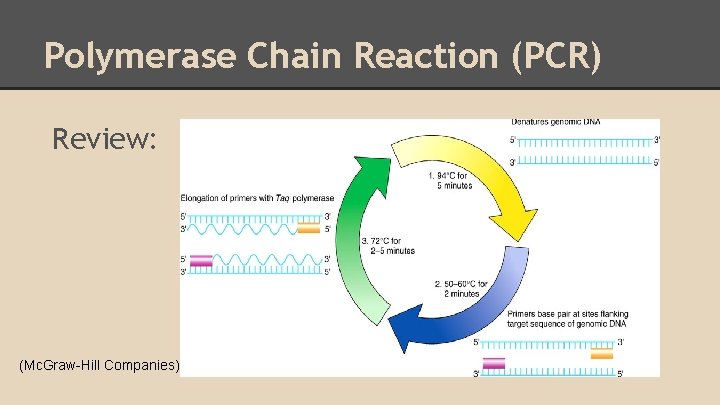
Polymerase Chain Reaction (PCR) Review: (Mc. Graw-Hill Companies)

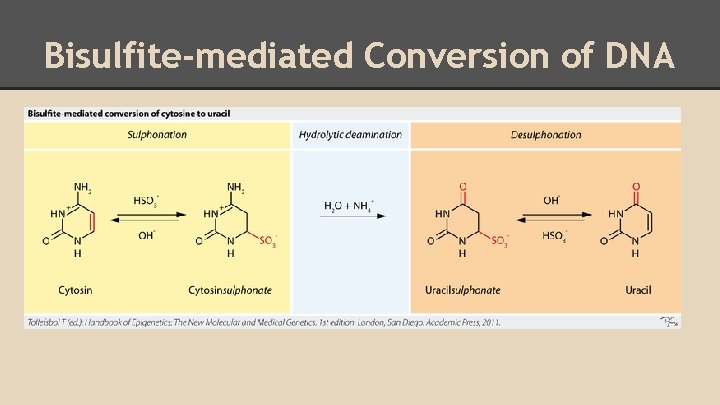
Bisulfite-mediated Conversion of DNA

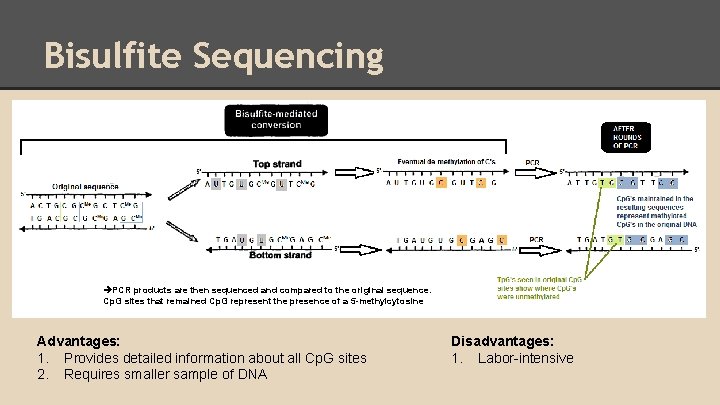
Bisulfite Sequencing PCR products are then sequenced and compared to the original sequence. Cp. G sites that remained Cp. G represent the presence of a 5 -methylcytosine Advantages: 1. Provides detailed information about all Cp. G sites 2. Requires smaller sample of DNA Disadvantages: 1. Labor-intensive

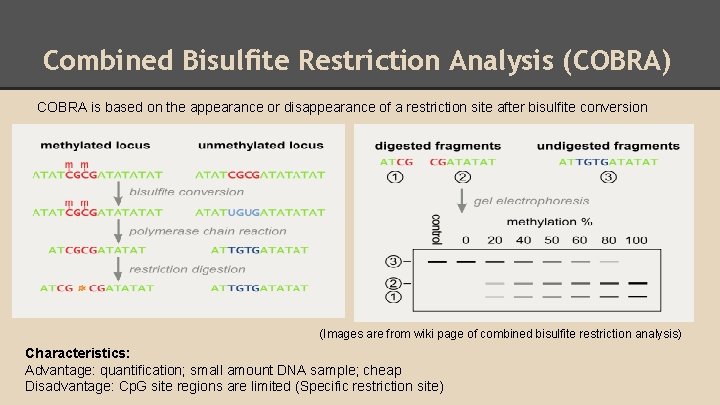
Combined Bisulfite Restriction Analysis (COBRA) COBRA is based on the appearance or disappearance of a restriction site after bisulfite conversion (Images are from wiki page of combined bisulfite restriction analysis) Characteristics: Advantage: quantification; small amount DNA sample; cheap Disadvantage: Cp. G site regions are limited (Specific restriction site)

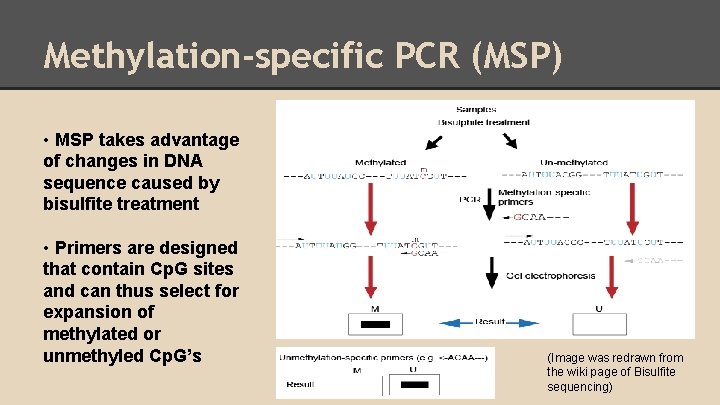
Methylation-specific PCR (MSP) • MSP takes advantage of changes in DNA sequence caused by bisulfite treatment • Primers are designed that contain Cp. G sites and can thus select for expansion of methylated or unmethyled Cp. G’s (Image was redrawn from the wiki page of Bisulfite sequencing)

MSP (cont’d) Advantages: Disadvantages: • Simple technique • Flexibility • Cheap • Optimal number of PCR cycles & annealing temperatures • Appropriate negative control • Not a quantification method

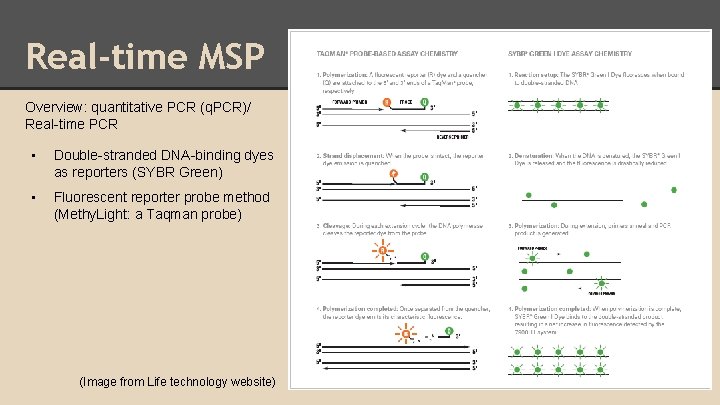
Real-time MSP Overview: quantitative PCR (q. PCR)/ Real-time PCR • Double-stranded DNA-binding dyes as reporters (SYBR Green) • Fluorescent reporter probe method (Methy. Light: a Taqman probe) (Image from Life technology website)

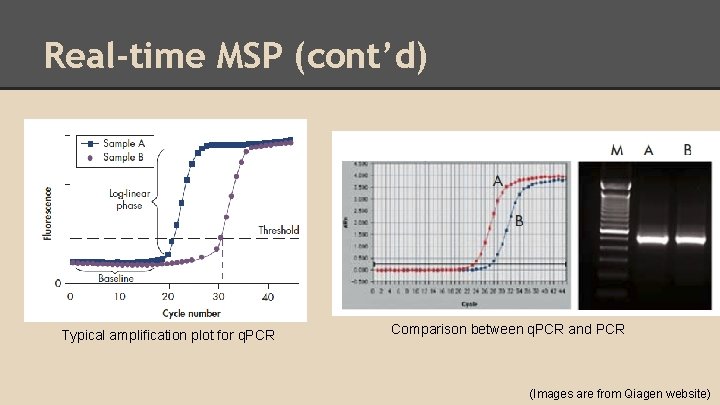
Real-time MSP (cont’d) Typical amplification plot for q. PCR Comparison between q. PCR and PCR (Images are from Qiagen website)

Real-time MSP (cont’d) Experiment Procedure: Bisulfite conversion; design methylation- and unmethylation-specific primers Results analysis: Comparing amplification of test samples with standard samples that contain known numbers of DNA molecules. Characteristics: High flexibility; accuracy; quantification; small amount samples; low cost Methylight has higher specificity than SYBR Green based real-time MSP

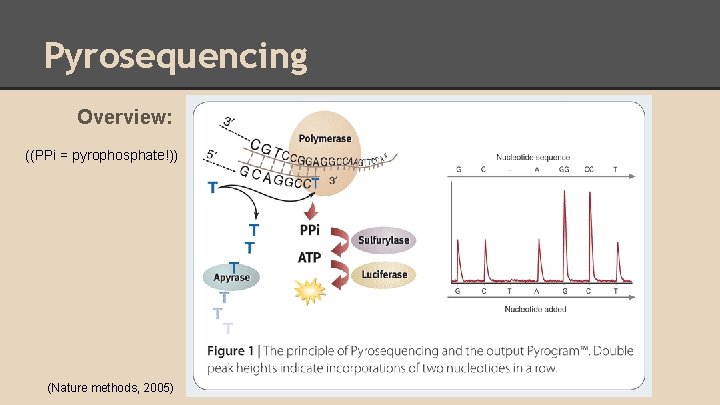
Pyrosequencing Overview: ((PPi = pyrophosphate!)) (Nature methods, 2005)

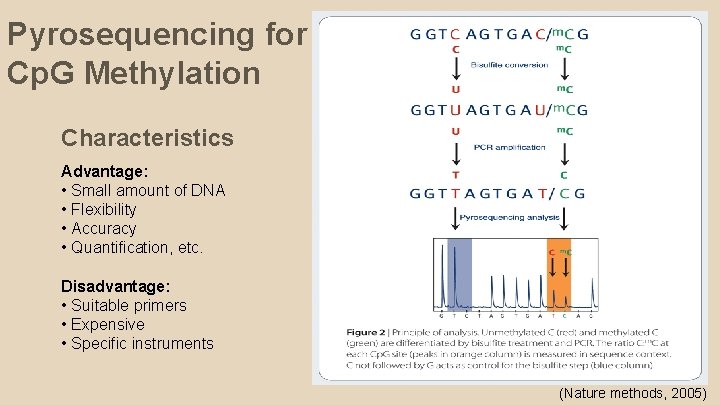
Pyrosequencing for Cp. G Methylation Characteristics Advantage: • Small amount of DNA • Flexibility • Accuracy • Quantification, etc. Disadvantage: • Suitable primers • Expensive • Specific instruments (Nature methods, 2005)

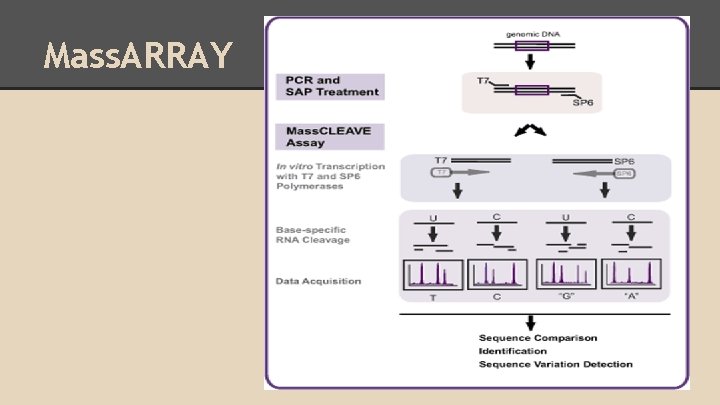
Mass. ARRAY

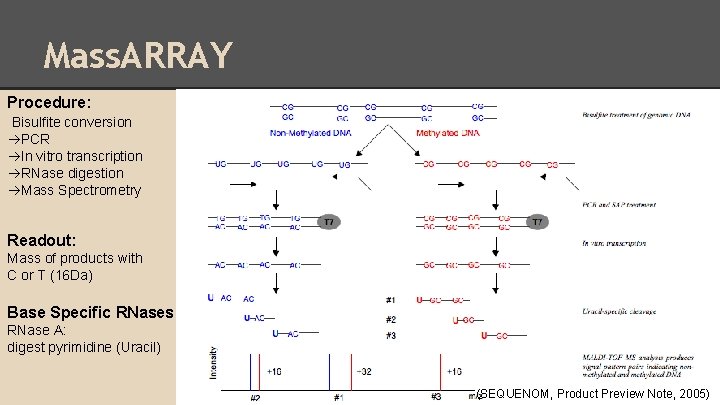
Mass. ARRAY Procedure: Bisulfite conversion PCR In vitro transcription RNase digestion Mass Spectrometry Readout: Mass of products with C or T (16 Da) Base Specific RNases RNase A: digest pyrimidine (Uracil) (SEQUENOM, Product Preview Note, 2005)

Mass. ARRAY Characteristics: Advantage: • Small amount of DNA • Flexibility • Quantification Disadvantage: • Expensive • Specific instruments

References Bird, A. P. , Southern, E. M. “Use of Restriction Enzymes to Study Eukaryotic DNA Methylation”. J. Mol. Biol. 118 (1978). 27 -37. Critical Factors for Successful Real-Time PCR. Qiagen, Real-Time PCR Brochure, 2010. Ehrich, M. et al. , Introduction to DNA Methylation Analysis Using the Mass. ARRAY System. SEQUENOM Preview Note, 2005. Herman, James G. “Methylation-specific PCR: A novel PCR assay for methylation status of Cp. G islands”. Proc, Natl. Acad. Sci. USA. 93 (1996). 9821 -9826. Pyro Q-Cp. GTM: quantitative analysis of methylation in multiple Cp. G sites by Pyrosequencing. Nature Methods 2005, 2. doi: 10. 1038/nmeth 800 Taq. Man® Chemistry vs. SYBR® Chemistry for Real-Time PCR. Life Technologies, q. PCR Education. Tollefsbol, Trygve. Handbook of Epigenetics: The New Molecular and Medical Genetics. Oxford, UK: Elsevier Inc. , 2011. 125 -134. Print. Wikipedia: Bisulfite sequencing http: //en. wikipedia. org/wiki/Bisulfite_sequencing Wikipedia: Combined bisulfite restriction analysis http: //en. wikipedia. org/wiki/Combined_bisulfite_restriction_analysis
 Massarray
Massarray Daj nam svima duha svog note
Daj nam svima duha svog note Palminervia
Palminervia Bắc cực
Bắc cực Lịch sử hà nội từ năm 1802 đến năm 1884
Lịch sử hà nội từ năm 1802 đến năm 1884 Khoi nguyen education group
Khoi nguyen education group Methylation vs acetylation
Methylation vs acetylation Methylation & chip-on-chip microarray platform
Methylation & chip-on-chip microarray platform Piperic acid to piperonal
Piperic acid to piperonal Adenine methylation
Adenine methylation Molecular ecological network analyses
Molecular ecological network analyses Critical thinking example in nursing
Critical thinking example in nursing Rhetorical analysis
Rhetorical analysis Courtage analyses services
Courtage analyses services When can their glory fade
When can their glory fade Facilitating learning child and adolescent development
Facilitating learning child and adolescent development Elements of sound in film
Elements of sound in film Microwave communication system
Microwave communication system Fade margin formula microwave
Fade margin formula microwave Tanulni kell az őszi fákat
Tanulni kell az őszi fákat Bursy fade
Bursy fade Window movi maker
Window movi maker Srop fade
Srop fade