Epigenetics DNA methylation II DNA methylation in plants



- Slides: 12

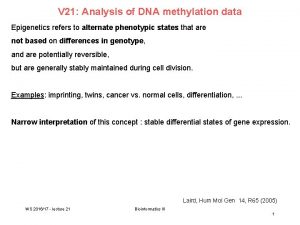
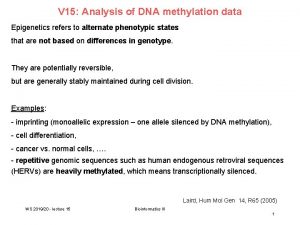
Epigenetics: DNA methylation II

DNA methylation in plants (Ch 9) Differences between plants and animals • Auxotroph vs. heterotroph • Sessile vs. mobile • Motionless and rigid vs. migrating and flexible cells Similarities • Chromatin structure and related machineries • Genome size and gene structures • DNA methylation (Cytosine)

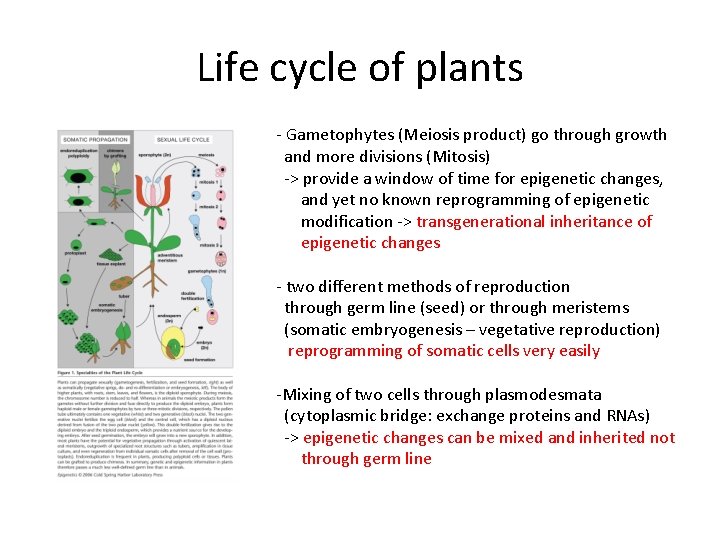
Life cycle of plants - Gametophytes (Meiosis product) go through growth and more divisions (Mitosis) -> provide a window of time for epigenetic changes, and yet no known reprogramming of epigenetic modification -> transgenerational inheritance of epigenetic changes - two different methods of reproduction through germ line (seed) or through meristems (somatic embryogenesis – vegetative reproduction) reprogramming of somatic cells very easily -Mixing of two cells through plasmodesmata (cytoplasmic bridge: exchange proteins and RNAs) -> epigenetic changes can be mixed and inherited not through germ line

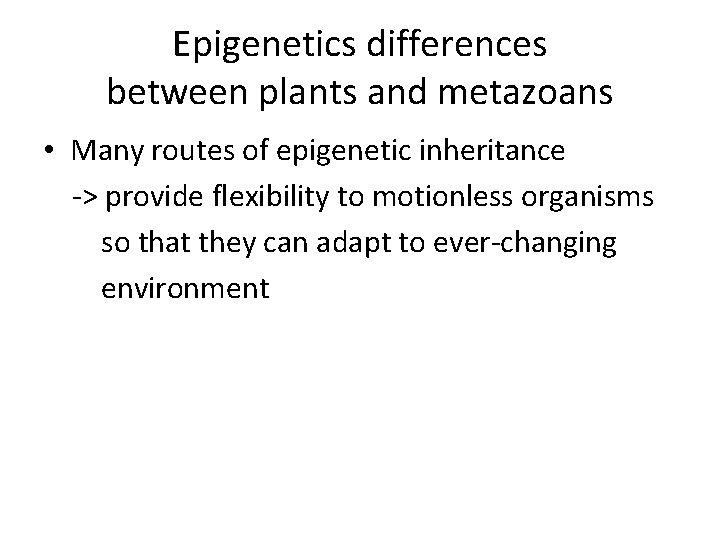
Epigenetics differences between plants and metazoans • Many routes of epigenetic inheritance -> provide flexibility to motionless organisms so that they can adapt to ever-changing environment

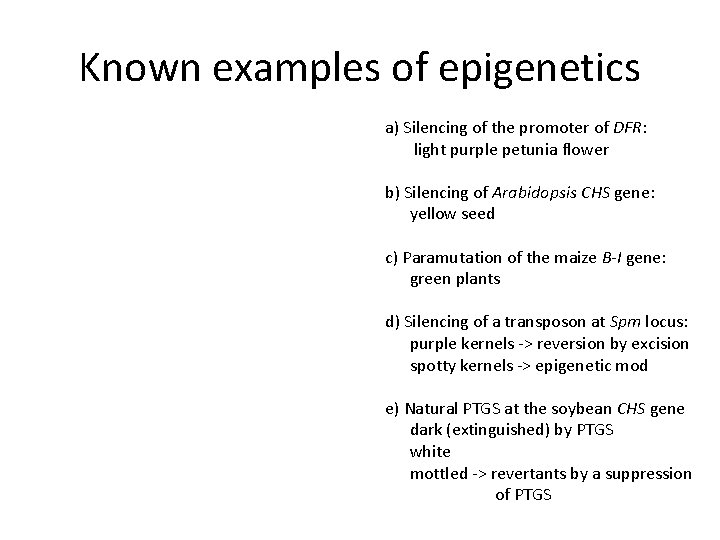
Known examples of epigenetics a) Silencing of the promoter of DFR: light purple petunia flower b) Silencing of Arabidopsis CHS gene: yellow seed c) Paramutation of the maize B-I gene: green plants d) Silencing of a transposon at Spm locus: purple kernels -> reversion by excision spotty kernels -> epigenetic mod e) Natural PTGS at the soybean CHS gene dark (extinguished) by PTGS white mottled -> revertants by a suppression of PTGS

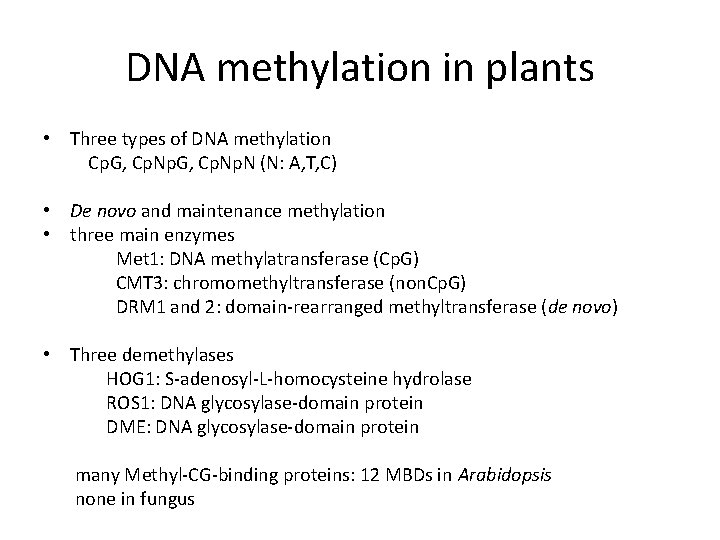
DNA methylation in plants • Three types of DNA methylation Cp. G, Cp. Np. N (N: A, T, C) • De novo and maintenance methylation • three main enzymes Met 1: DNA methylatransferase (Cp. G) CMT 3: chromomethyltransferase (non. Cp. G) DRM 1 and 2: domain-rearranged methyltransferase (de novo) • Three demethylases HOG 1: S-adenosyl-L-homocysteine hydrolase ROS 1: DNA glycosylase-domain protein DME: DNA glycosylase-domain protein many Methyl-CG-binding proteins: 12 MBDs in Arabidopsis none in fungus

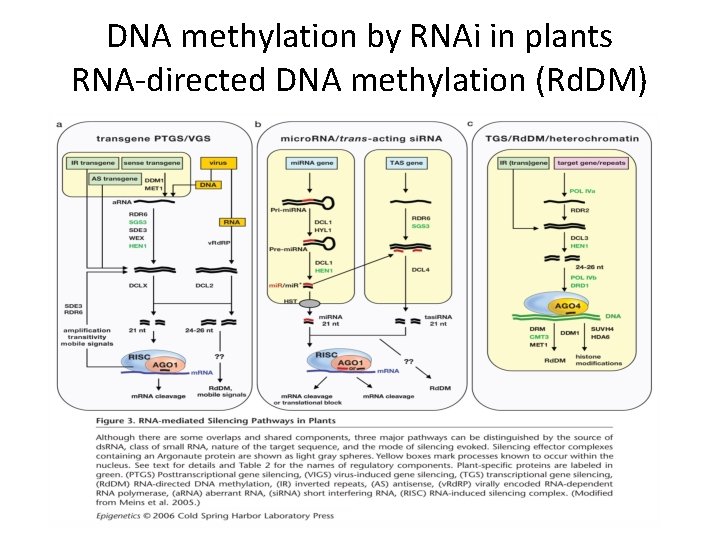
DNA methylation by RNAi in plants RNA-directed DNA methylation (Rd. DM)

DNA methylation in fungi (Neurospora crassa) - limited DNA methylation (1. 5% Cytosines) r. DNA and repeats - part of Repeat-induced point mutation (RIP) - defense mechanism - only one enzyme DMT-2, but no other machineries for recognition (MBDs)

Major questions • What triggers DNA methylation? (histone connections) • What protects Cp. G islands from DNA methylation? • How DNA methylation maintained in somatic cells (how DNMT 1 transcript and protein are controlled? )

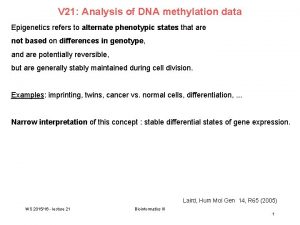
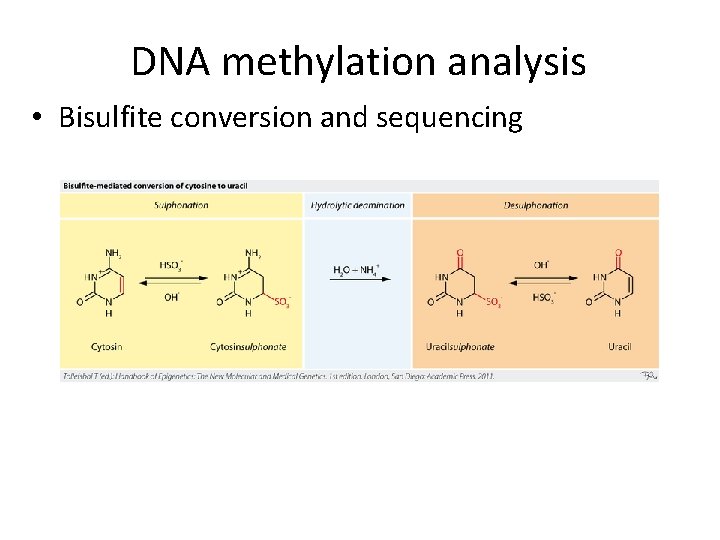
DNA methylation analysis • Bisulfite conversion and sequencing

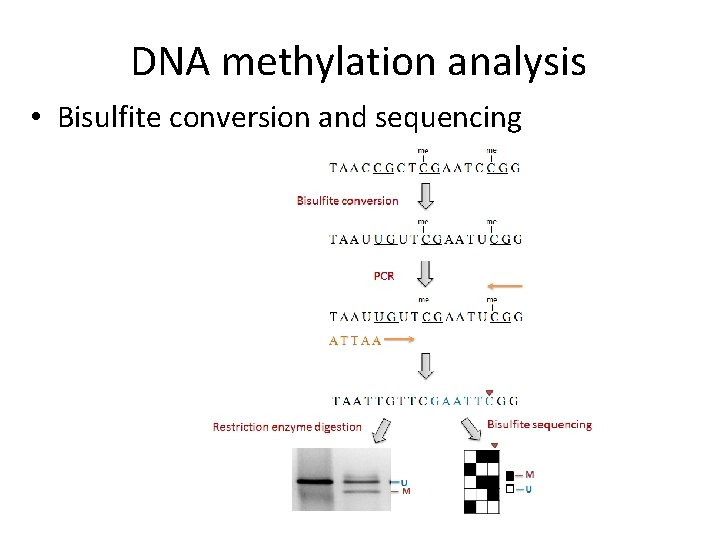
DNA methylation analysis • Bisulfite conversion and sequencing

Papers to be discussed • Sept 4 th : Transgenerational inheritance of stress through small non-coding RNAs Gapp K et al. Nature Neuroscience 2014.
 Histone modification epigenetics
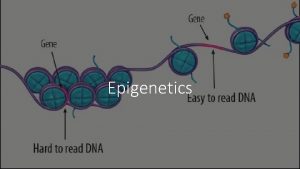
Histone modification epigenetics What is epigenetics
What is epigenetics Methylation vs acetylation
Methylation vs acetylation Methylation & chip-on-chip microarray platform
Methylation & chip-on-chip microarray platform Hofmann exhaustive methylation
Hofmann exhaustive methylation Adenine methylation
Adenine methylation Vascular plants vs nonvascular plants
Vascular plants vs nonvascular plants Non vascular plants
Non vascular plants Characteristics of flowering and non flowering plants
Characteristics of flowering and non flowering plants Photosynthesis equation
Photosynthesis equation What role does dna polymerase play in copying dna?
What role does dna polymerase play in copying dna? Replication fork
Replication fork Chapter 11 dna and genes
Chapter 11 dna and genes