EPIGENETICS IN CANCER THERAPY DNA Methylation Methylated DNA


- Slides: 12

EPIGENETICS IN CANCER THERAPY

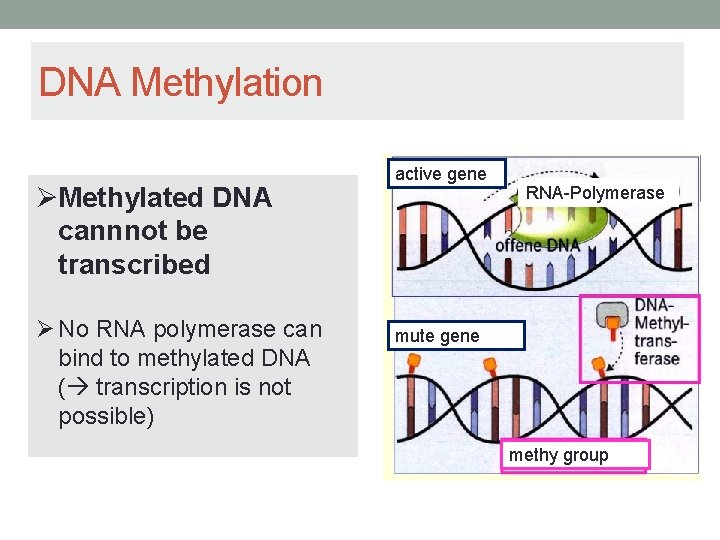
DNA Methylation Methylated DNA cannnot be transcribed No RNA polymerase can bind to methylated DNA ( transcription is not possible) active gene RNA-Polymerase mute gene methy group

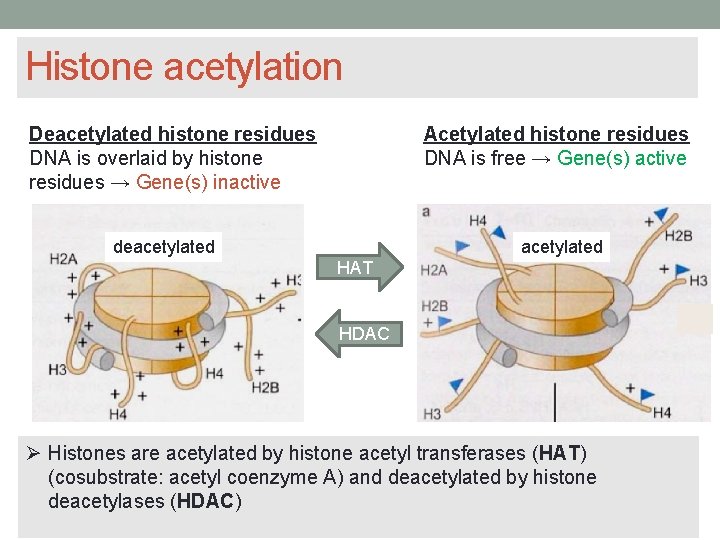
Histone acetylation Acetylated histone residues DNA is free → Gene(s) active Deacetylated histone residues DNA is overlaid by histone residues → Gene(s) inactive deacetylated HAT HDAC Histones are acetylated by histone acetyl transferases (HAT) (cosubstrate: acetyl coenzyme A) and deacetylated by histone deacetylases (HDAC)

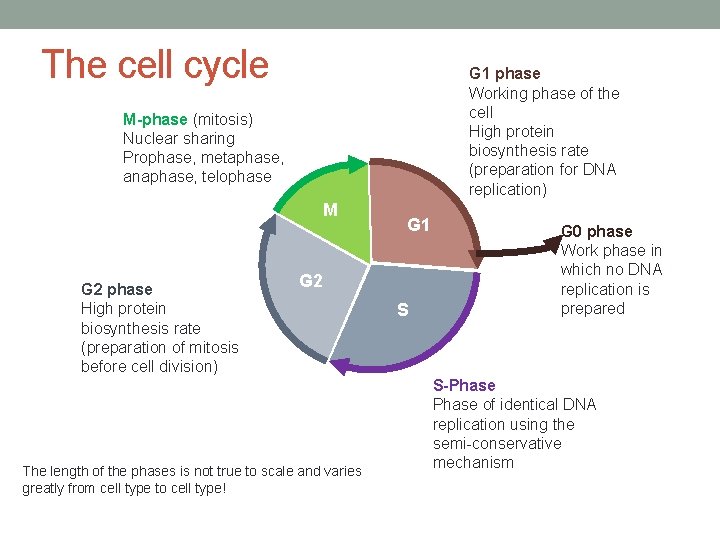
The cell cycle G 1 phase Working phase of the cell High protein biosynthesis rate (preparation for DNA replication) M-phase (mitosis) Nuclear sharing Prophase, metaphase, anaphase, telophase M G 2 phase High protein biosynthesis rate (preparation of mitosis before cell division) G 1 G 2 The length of the phases is not true to scale and varies greatly from cell type to cell type! S G 0 phase Work phase in which no DNA replication is prepared S-Phase of identical DNA replication using the semi-conservative mechanism

Genetic regulation of cell growth Cell growth regulating genes are grouped into two types: 1. Tumor suppressor genes and their expressed gene products have to slow down cell cycle 2. Proto-oncogenes and their gene products have to accellerate the cell cycle Tumor-suppressor genes and proto-oncogenes are working together in an equilibration regulating the speed of cell cycle in a perfect way, You can compare this genetic growth regulation with speed regulation in a car. Both the accelerator and the brake are neccessary to regulate the speed of a car.

Epigenetic changes in tumor cells The tumor suppressor proteins investigated so far slow down the cell cycle But DNA of tumor cells within the regulatory sequences of tumor suppressor genes is highly methylated TSP genes are inactive Gene products of tumor suppressor genes are not produced by inactive TSP

Epigenetic changes in tumor cells Retinoblastoma is a tumor on the retina inside the eye. Retinoblastoma gene is a tumor suppressor gene o It has like each genes two allels located on chromosome 13 o If both allels are mutated the retina cell gets maligne and the cell is growing to a tumor Expressed Retinoblastoma protein prevents the transition from the G 1 to the S phase of the cell cycle → If the retinoblastoma protein is not expressed by methylation, a tumor cell starts to divide very fast

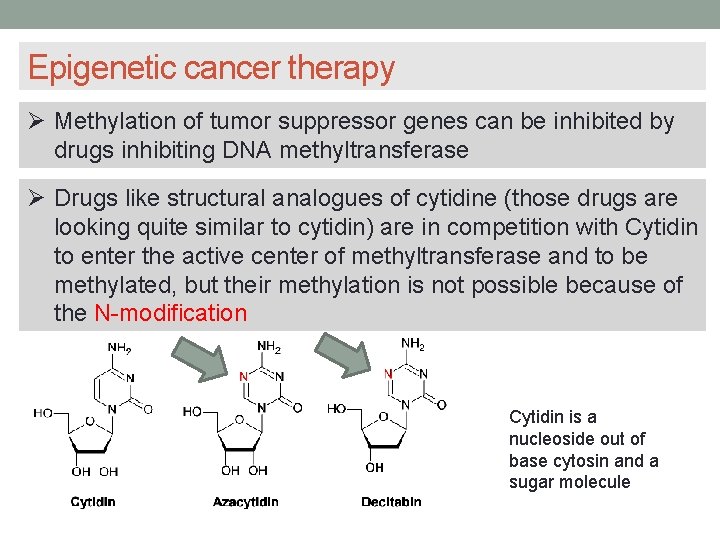
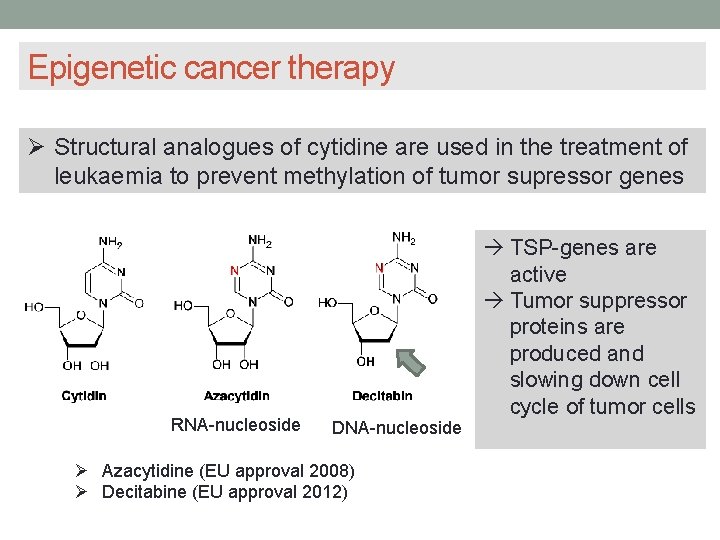
Epigenetic cancer therapy Methylation of tumor suppressor genes can be inhibited by drugs inhibiting DNA methyltransferase Drugs like structural analogues of cytidine (those drugs are looking quite similar to cytidin) are in competition with Cytidin to enter the active center of methyltransferase and to be methylated, but their methylation is not possible because of the N-modification Cytidin is a nucleoside out of base cytosin and a sugar molecule

Epigenetic cancer therapy Structural analogues of cytidine are used in the treatment of leukaemia to prevent methylation of tumor supressor genes RNA-nucleoside DNA-nucleoside Azacytidine (EU approval 2008) Decitabine (EU approval 2012) TSP-genes are active Tumor suppressor proteins are produced and slowing down cell cycle of tumor cells

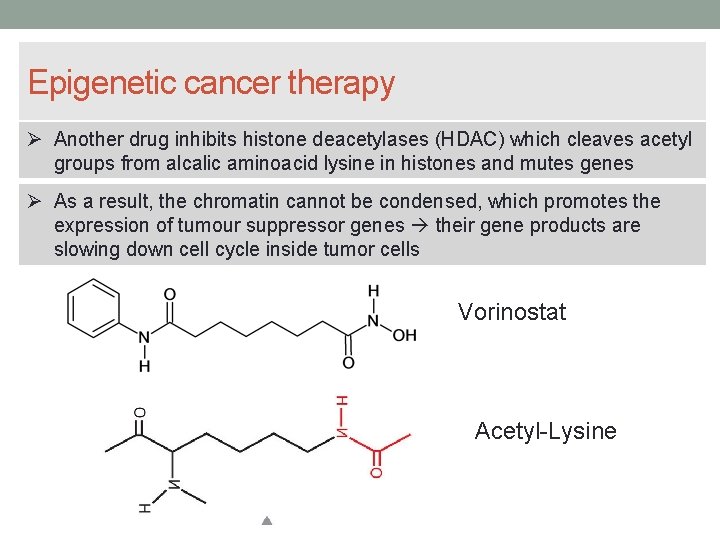
Epigenetic cancer therapy Another drug inhibits histone deacetylases (HDAC) which cleaves acetyl groups from alcalic aminoacid lysine in histones and mutes genes As a result, the chromatin cannot be condensed, which promotes the expression of tumour suppressor genes their gene products are slowing down cell cycle inside tumor cells Vorinostat Acetyl-Lysine

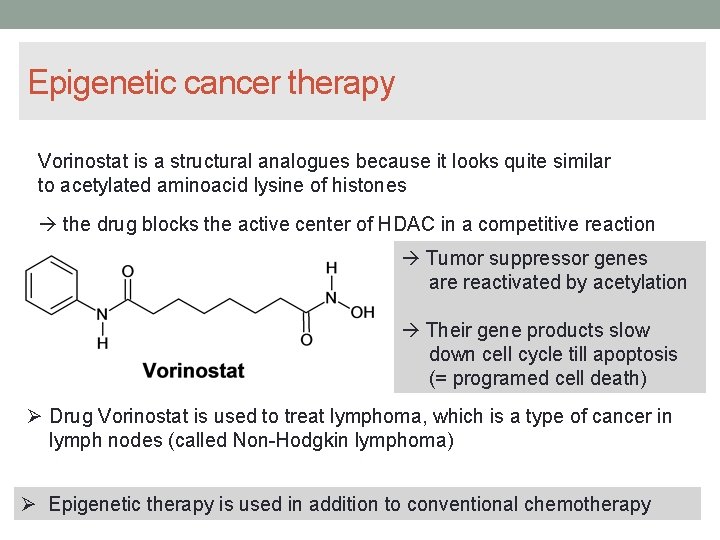
Epigenetic cancer therapy Vorinostat is a structural analogues because it looks quite similar to acetylated aminoacid lysine of histones the drug blocks the active center of HDAC in a competitive reaction Tumor suppressor genes are reactivated by acetylation Their gene products slow down cell cycle till apoptosis (= programed cell death) Drug Vorinostat is used to treat lymphoma, which is a type of cancer in lymph nodes (called Non-Hodgkin lymphoma) Epigenetic therapy is used in addition to conventional chemotherapy

Conclusions of this presentation 1. Drugs for cancer treatment working on the level of epigenetic are still on the market and a huge benefit in cancer therapy for many patients. 2. Molecular working mechanism of those drugs is as structural analogues of enzymes regulating gene activity on epigenetic level.
 Can you use methylated spirits in a trangia?
Can you use methylated spirits in a trangia? Adenine methylation
Adenine methylation Herzig meyer method
Herzig meyer method Early fly
Early fly Methylation vs acetylation
Methylation vs acetylation What is epigenetics
What is epigenetics Methylation & chip-on-chip microarray platform
Methylation & chip-on-chip microarray platform Proton therapy for breast cancer after mastectomy
Proton therapy for breast cancer after mastectomy Psychoanalytic therapy is to as humanistic therapy is to
Psychoanalytic therapy is to as humanistic therapy is to Psychoanalytic therapy is to as humanistic therapy is to
Psychoanalytic therapy is to as humanistic therapy is to Bioness integrated therapy system occupational therapy
Bioness integrated therapy system occupational therapy Elite dna therapy services llc
Elite dna therapy services llc What role does dna polymerase play in copying dna?
What role does dna polymerase play in copying dna?