Coregistration and Spatial Normalisation Martin Chadwick and Catherine








- Slides: 26

Co-registration and Spatial Normalisation Martin Chadwick and Catherine Sebastian

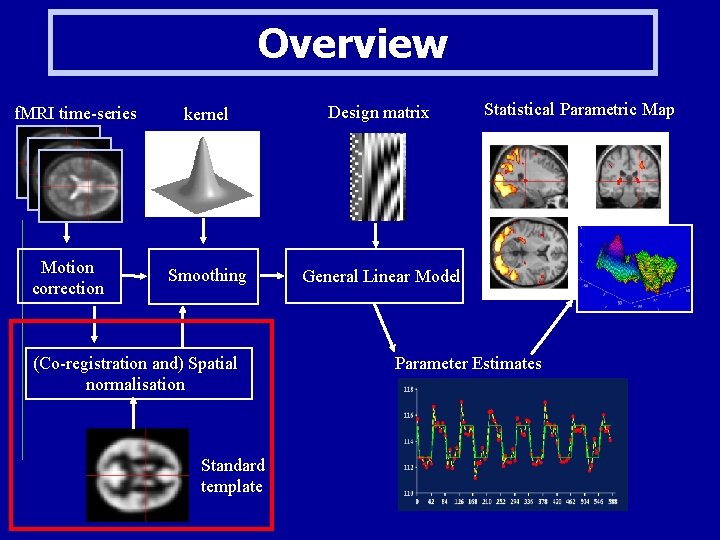
Overview f. MRI time-series Motion correction kernel Design matrix Smoothing General Linear Model (Co-registration and) Spatial normalisation Standard template Statistical Parametric Map Parameter Estimates

Preprocessing Steps • Realignment – Motion correction: Adjust for movement between slices • Coregistration – Overlay structural and functional images: Link functional scans to anatomical scan • Normalisation – Warp images to fit to a standard template brain • Smoothing – To increase signal-to-noise ratio • Extras (optional) – Slice timing correction; unwarping

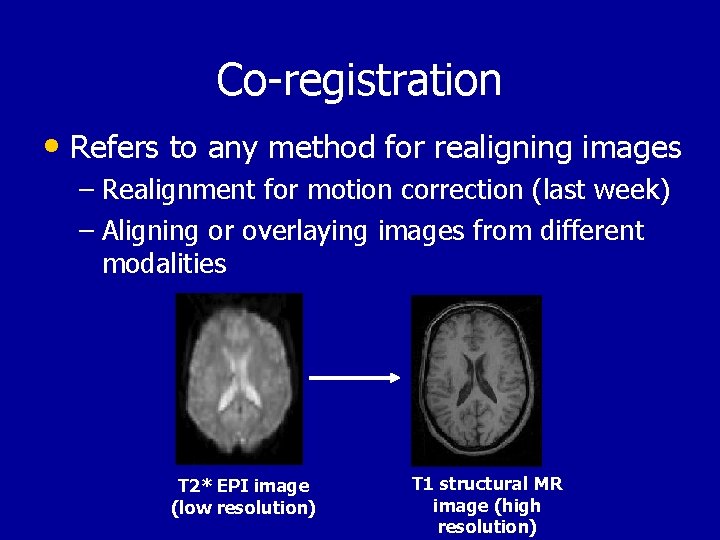
Co-registration • Refers to any method for realigning images – Realignment for motion correction (last week) – Aligning or overlaying images from different modalities T 2* EPI image (low resolution) T 1 structural MR image (high resolution)

Why Co-register structural and functional images? • Can overlay functional activations onto an individual’s own anatomy • Can overlay group-level functional activations onto an average structural • Gives you a better spatial image for later normalisation step, as warps derived from the higher resolution structural image can be applied to the functional image

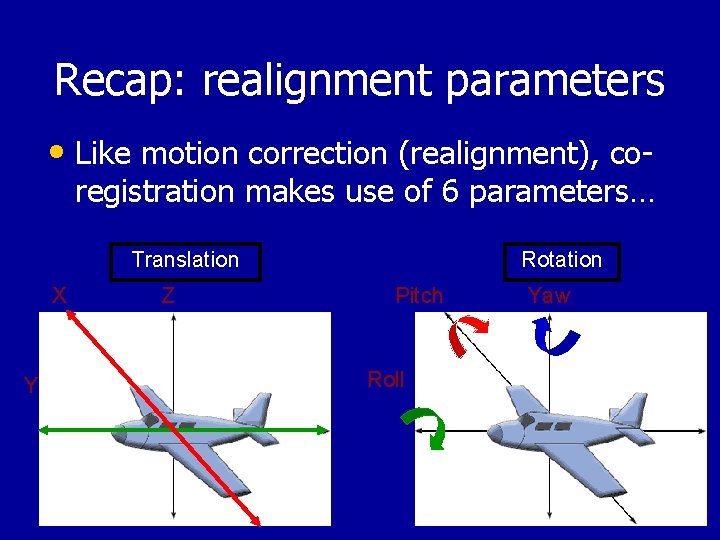
Recap: realignment parameters • Like motion correction (realignment), co- registration makes use of 6 parameters… Translation X Y Z Rotation Pitch Roll Yaw

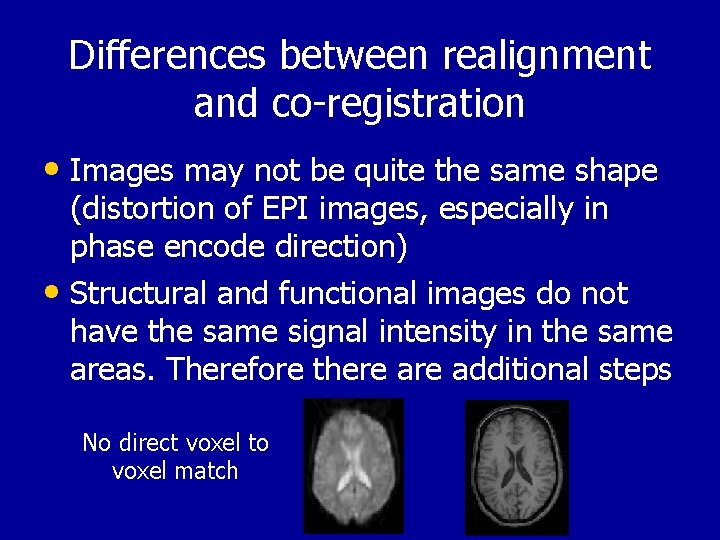
Differences between realignment and co-registration • Images may not be quite the same shape (distortion of EPI images, especially in phase encode direction) • Structural and functional images do not have the same signal intensity in the same areas. Therefore there additional steps No direct voxel to voxel match

The Normalised Mutual Information Approach • Different material will have different intensities within a scan modality (e. g. air will have a consistent brightness, and this will differ from other materials such as white matter). • When looking between modalities, these consistencies can be used to compare images

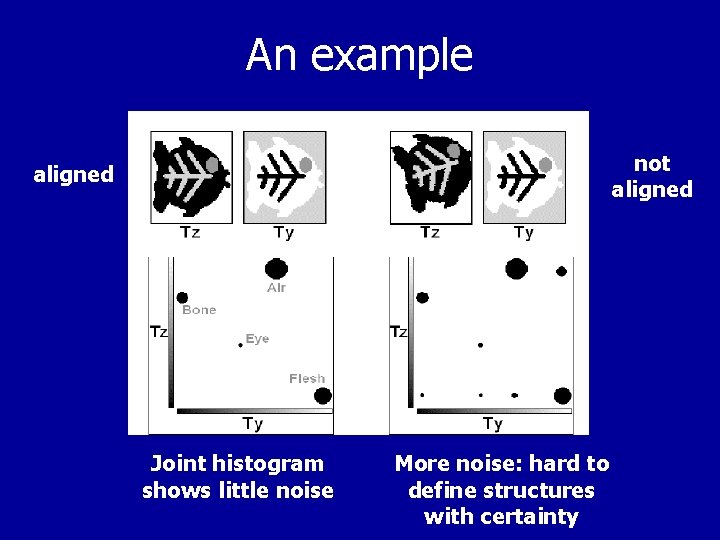
An example not aligned Joint histogram shows little noise More noise: hard to define structures with certainty

Additional points • In some studies structurals are not taken – it is possible to conduct f. MRI analysis without co-registering to a structural • Sometimes when you co-register, you have to reslice the data - E. g. change image dimensions from 3 x 3 x 3 to 2 x 2 x 2, or change apparent direction of data collection from axial to coronal - Useful if two images have very different voxel sizes - Often involves interpolation - Often used with PET data

Co-registration in SPM

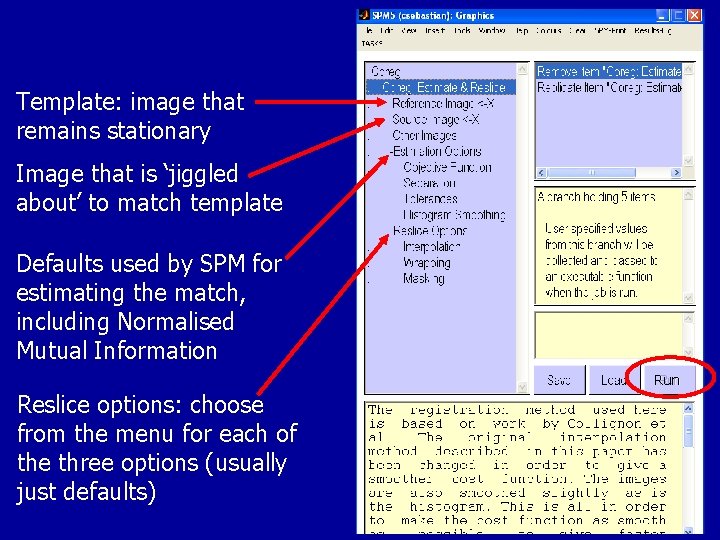
Co-registration in SPM Make selection Explains each option

Template: image that remains stationary Image that is ‘jiggled about’ to match template Defaults used by SPM for estimating the match, including Normalised Mutual Information Run Reslice options: choose from the menu for each of the three options (usually just defaults)

Preprocessing Steps • Realignment – Motion correction: Adjust for movement between slices • Coregistration – Overlay structural and functional images: Link functional scans to anatomical scan • Normalisation – Warp images to fit to a standard template brain • Smoothing – To increase signal-to-noise ratio • Extras (optional) – Slice timing correction; unwarping

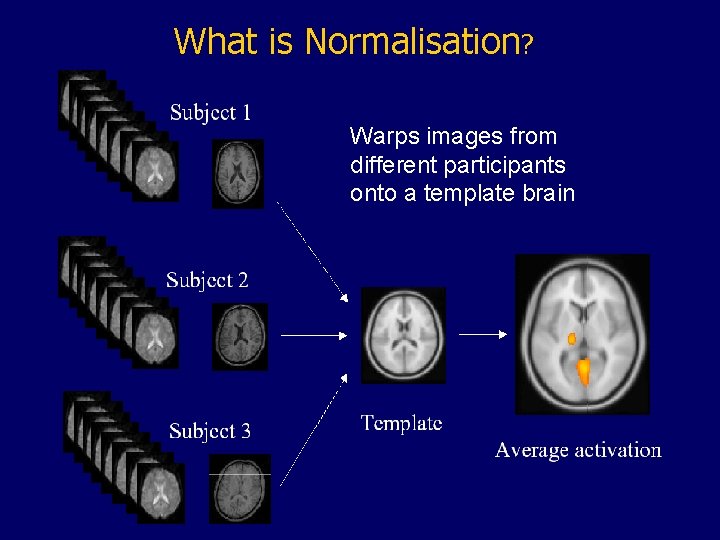
What is Normalisation? Warps images from different participants onto a template brain Matthew Brett

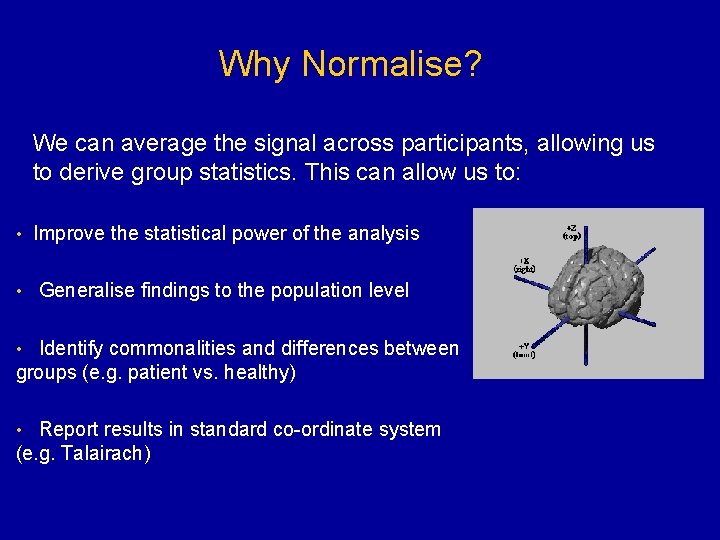
Why Normalise? We can average the signal across participants, allowing us to derive group statistics. This can allow us to: • Improve the statistical power of the analysis • Generalise findings to the population level • Identify commonalities and differences between groups (e. g. patient vs. healthy) • Report results in standard co-ordinate system (e. g. Talairach)

SPM: Spatial Normalisation • SPM uses a voxel-intensity-based approach to normalisation. • adopts a two-stage procedure : • Step 1: Linear transformation (12 -parameter affine). This step accounts for the major differences in head shape and position, but there will be remaining smaller-scale differences. • Step 2: Non-linear transformation (warping). The non-linear step is designed to take care of the smaller-scale differences in brain anatomy. Alternatives – anatomy based approaches e. g. Free. Surfer

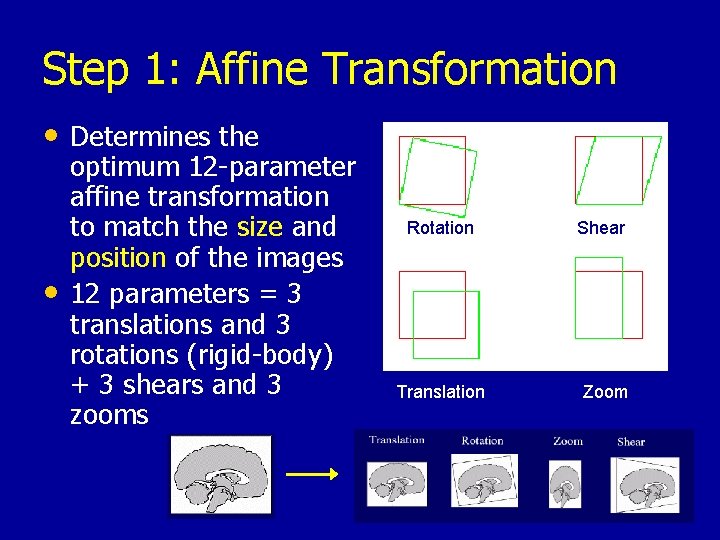
Step 1: Affine Transformation • Determines the • optimum 12 -parameter affine transformation to match the size and position of the images 12 parameters = 3 translations and 3 rotations (rigid-body) + 3 shears and 3 zooms Rotation Shear Translation Zoom

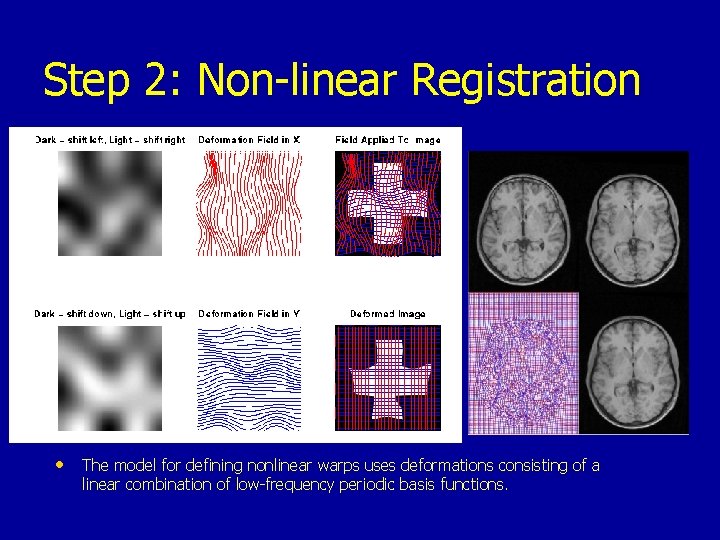
Step 2: Non-linear Registration • The model for defining nonlinear warps uses deformations consisting of a linear combination of low-frequency periodic basis functions.

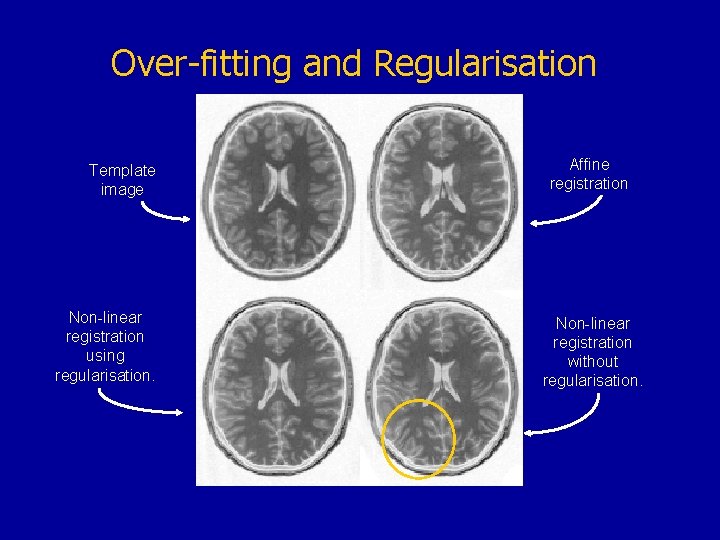
Over-fitting and Regularisation Template image Non-linear registration using regularisation. Affine registration Non-linear registration without regularisation.

Caveats • Impossible to make a meaningful perfect structural match between subjects, due to individual differences in anatomy • Even if anatomy is well-matched, it does not guarantee that functionally homologous areas are spatially aligned – we don’t know the extent to which individuals may vary in their structure-function relationships. • Pathology creates a particular problem here, as even relatively confined abnormalities or lesions can cause mis-registration in widespread areas of the brain, due to the global nature of the normalisation process. Need to bare this in mind in patient studies. Solution • A partial solution for any remaining small-scale differences in anatomical or functional location is offered by the next stage of pre-processing, where the images are spatially smoothed.

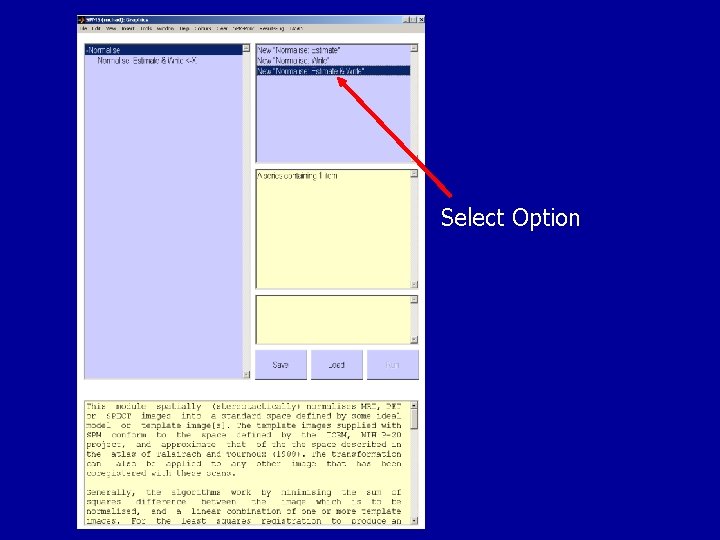
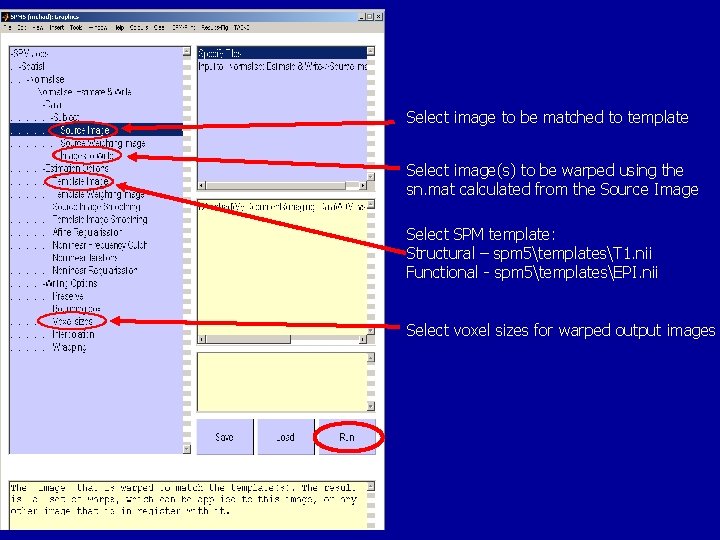
Normalisation in SPM

Select Option

Select image to be matched to template Select image(s) to be warped using the sn. mat calculated from the Source Image Select SPM template: Structural – spm 5templatesT 1. nii Functional - spm 5templatesEPI. nii Select voxel sizes for warped output images


Sources: • Friston, K. J. Introduction: Experimental design and statistical parametric mapping http: //www. fil. ion. ucl. ac. uk/spm/doc/intro/ • Ashburner & Friston “Rigid Body Registration” Chapter 2, Human Brain Function, 2 nd ed. ; http: //www. fil. ion. ucl. ac. uk/spm/doc/books/hbf 2/ • Ashburner & Friston “Spatial Normalization Using Basis Functions” Chapter 3, Human Brain Function, 2 nd ed. ; http: //www. fil. ion. ucl. ac. uk/spm/doc/books/hbf 2/ • Rik Henson’s Preprocessing Slides: http: //imaging. mrc-cbu. cam. ac. uk/imaging/Processing. Stream • Previous Mf. D Slides