Ch IPseq data analysis and visualization using Chipster




- Slides: 14

Ch. IP-seq data analysis and visualization using Chipster Workshop on next generation sequencing data analysis 31. 5 - 4. 6. 2010 Espoo Massimiliano Gentile CSC – IT Center for Science

Chipster What is it? User-friendly analysis software and workflow tool • Intuitive GUI, interactive visualizations • Analysis steps taken can be saved as an automatic workflow, which can be shared Generic platform • currently used mainly for microarray data and proteomics data • building support for Ch. IP-seq, RNA-seq and mi. RNA-seq Client-server system: centralized maintenance and updates • Also Web services (SOAP) are connected to the system Open source, server installation packages available • http: //chipster. sourceforge. net/ http: //chipster. csc. fi

Chipster Goals Enable researchers without programming skills or extensive bioinformatics knowledge to: • access to an extensive selection of up-to-date tools for highthroughput data analysis • work with the data through a graphical and intuitive user interface • combine tools into automatic workflows that can be shared • integrate different types of data and analysis workflows • interpret results in meaningful and efficient visualizations

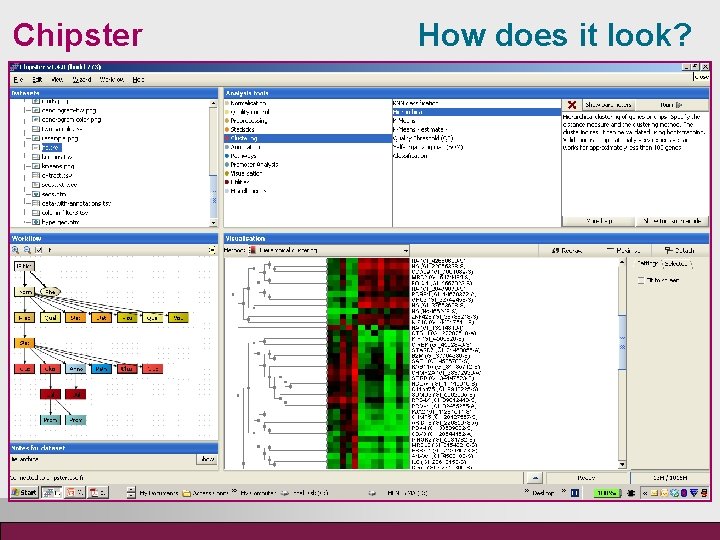
Chipster How does it look?

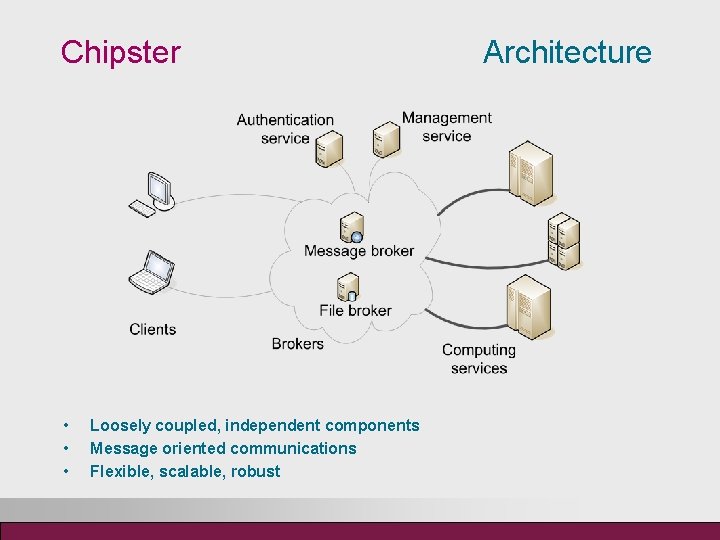
Chipster • • • Loosely coupled, independent components Message oriented communications Flexible, scalable, robust Architecture

Chipster NGS data analysis Currently building support for: Ch. IP-seq RNA-seq, mi. RNA-seq Me. DIP-seq, BS-seq Tools Preprocessing (merging, sorting, filtering, …) Alignment (Maq, Bowtie, Top. Hat, …) Peak detection (MACS, Peak. Seq, …) Motif and TFBS detection Finding neighbouring genes Pathway analysis RNA-seq: quantitation and detection of novel splice variants Integration with target gene expression Visualization Genome Browser

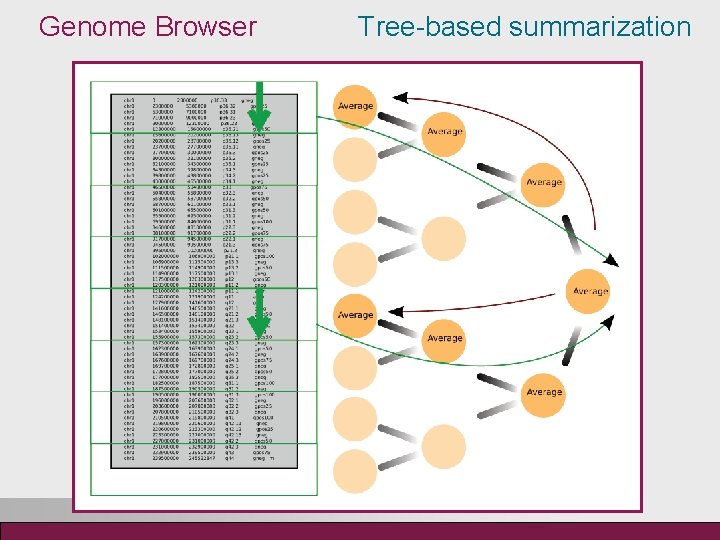
Genome Browser Features • Open source, java-based • Interactive zooming from full chromosome down to nucleotide level • Ensembl annotations for transcripts and genes including mi. RNA • Easily extendable with new tracks, views and file formats • Standalone as well as Integrated with Chipster analysis environment Challenges • Handle very large data sets • View both the big picture and the details • Smooth zooming and browsing Solution • Optimize global viewing by data sampling: details not read when looking at the big picture • Optimize local viewing: the whole data not read when looking at a detail • Both optimizations need random access to data, at the moment local files

Genome Browser Tree-based summarization

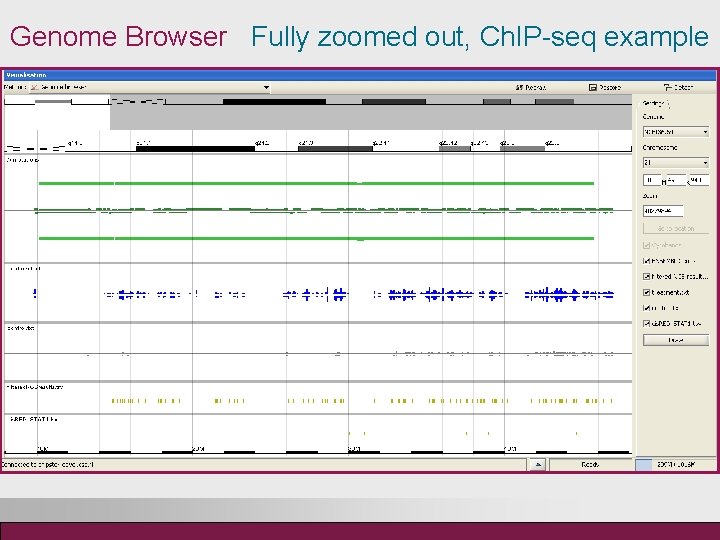
Genome Browser Fully zoomed out, Ch. IP-seq example

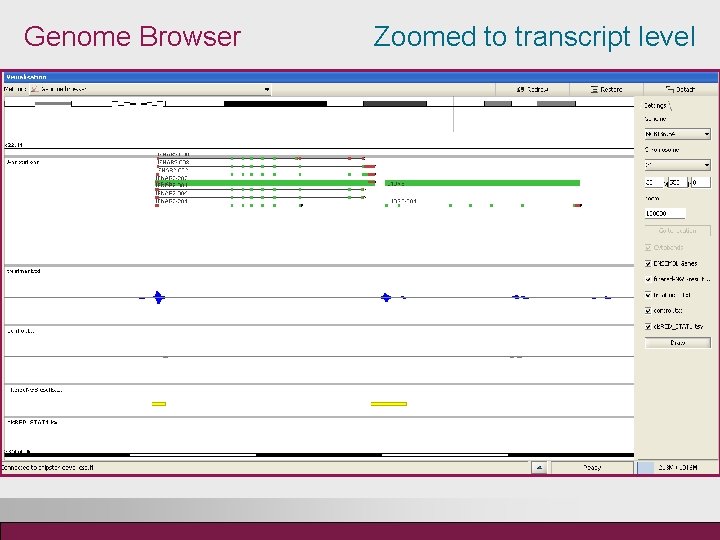
Genome Browser Zoomed to transcript level

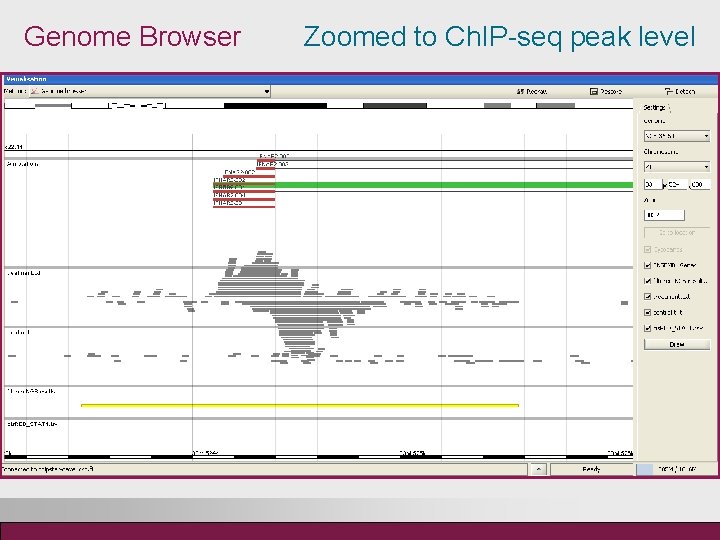
Genome Browser Zoomed to Ch. IP-seq peak level

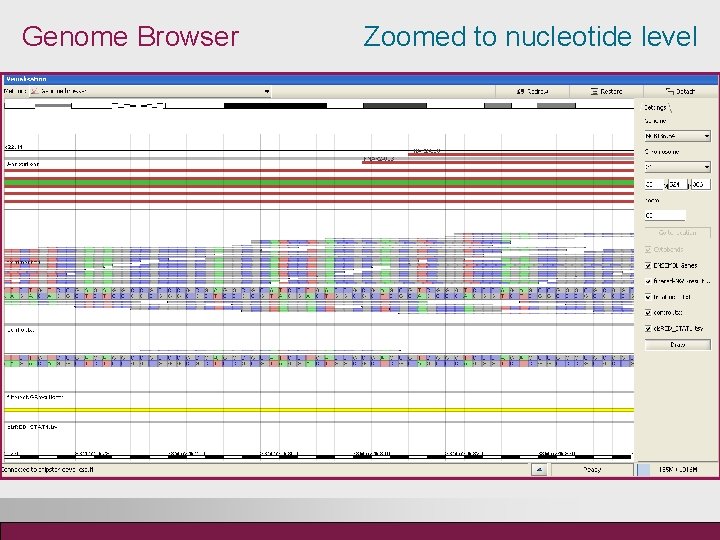
Genome Browser Zoomed to nucleotide level

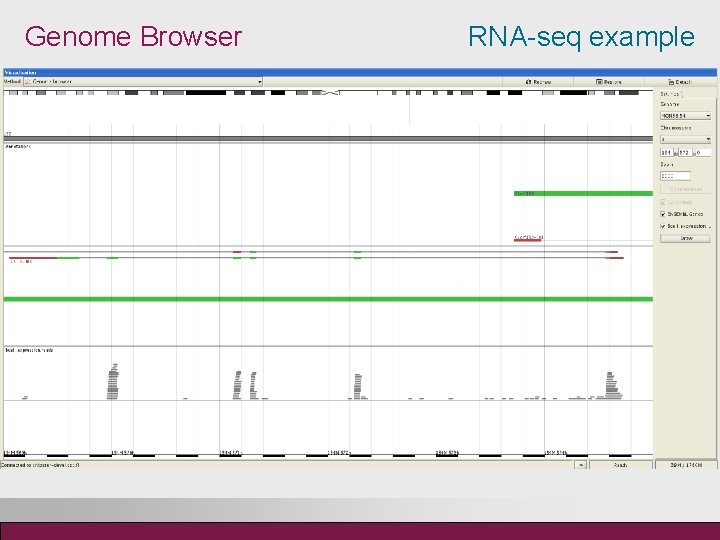
Genome Browser RNA-seq example

Acknowledgements Chipster development team Jarno Tuimala Eija Korpelainen Aleksi Kallio Taavi Hupponen Petri Klemelä Mikko Koski Janne Käki Collaborators Ilari Scheinin Laura Elo Dario Greco Funding agents Tekes (SYSBIO research programme) European Commission (FP 6 No. E EMBRACE)