Analysis of Affymetrix expression data using R on







- Slides: 18

Analysis of Affymetrix expression data using R on Azure Cloud Anne Owen Department of Mathematical Sciences University of Essex Dr Andrew Harrison, University of Essex Dr Hugh Shanahan, Royal Holloway, University of London SAICG Workshop, Oxford 15/16 March, 2012

Introduction • • • The Affymetrix Gene. Chip Micro-array data Venus-C pilot project R scripts on Azure Cloud Results to date Our Experience

• We are developing informatics tools to aid the analysis of Affymetrix chips (Gene. Chips, Exon arrays). • Micro-arrays are the data read from Gene. Chips Affymetrix Gene. Chip • Array. Express is an example of a public database containing microarrays and other data from biological experiments

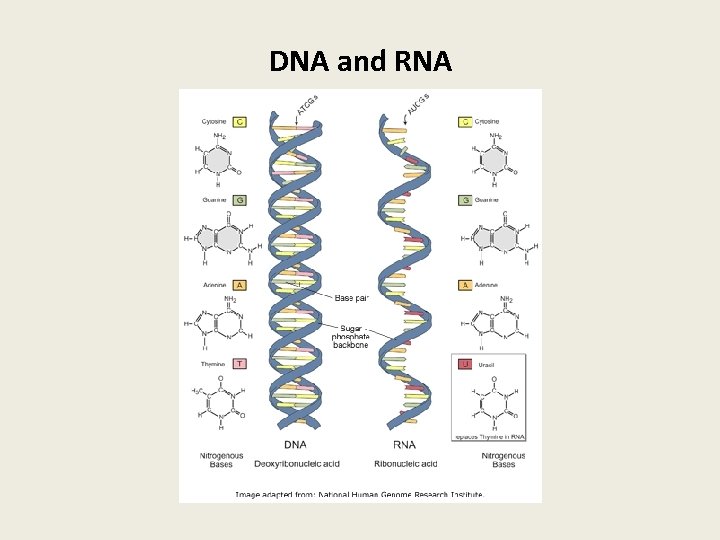
DNA and RNA

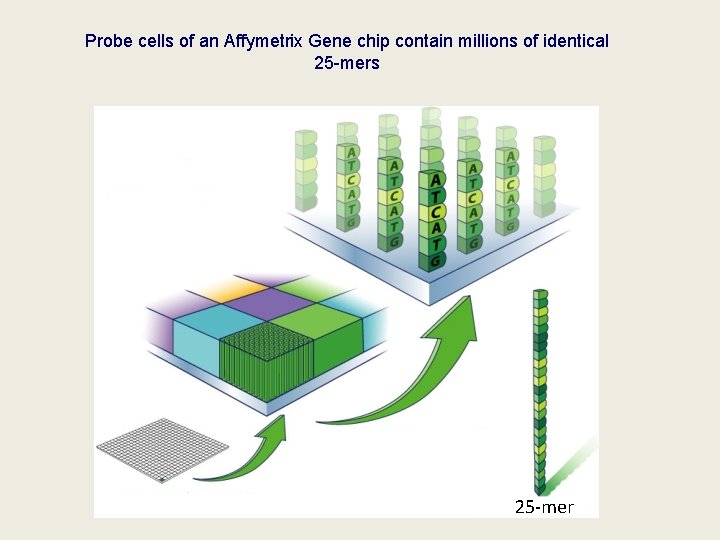
Probe cells of an Affymetrix Gene chip contain millions of identical 25 -mers 25 -mer

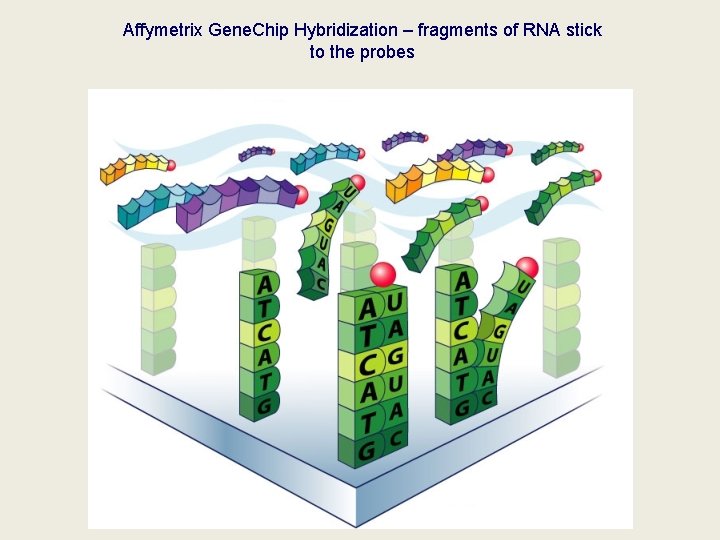
Affymetrix Gene. Chip Hybridization – fragments of RNA stick to the probes

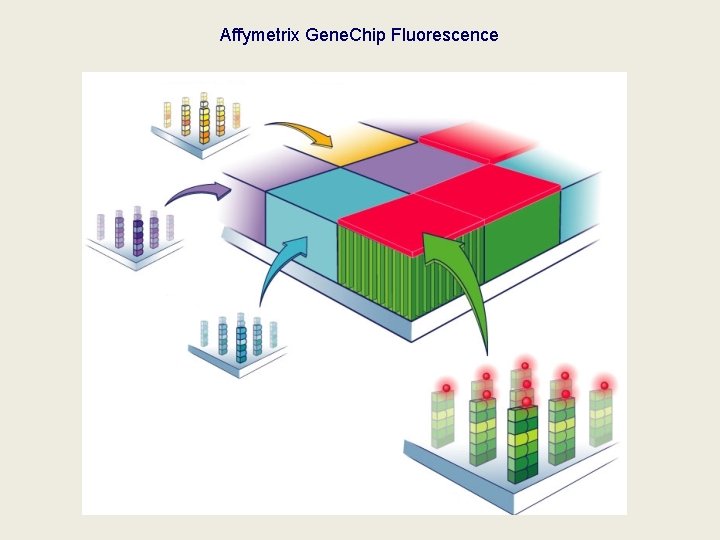
Affymetrix Gene. Chip Fluorescence

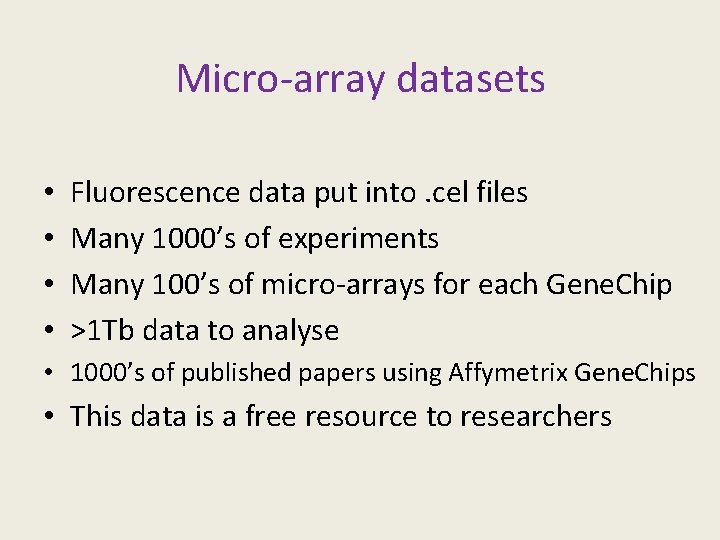
Micro-array datasets • • Fluorescence data put into. cel files Many 1000’s of experiments Many 100’s of micro-arrays for each Gene. Chip >1 Tb data to analyse • 1000’s of published papers using Affymetrix Gene. Chips • This data is a free resource to researchers

Going Forward. . . • Currently we analyse flaws in Genechip data • Future is new genomic technology known as ‘next generation sequencing’ • Petabytes of data being generated faster than it can be analysed • Cloud solutions needed for storage of and access to this data

Venus-C Pilot Project • VENUS-C is a project funded under the European Commission’s 7 th Framework Programme with computing resources from Microsoft • Joint co-operation between computing service providers and scientific user communities • Aim: to develop, test and deploy a large, Cloud computing infrastructure for science and SMEs (small and medium-sized enterprises) in Europe.


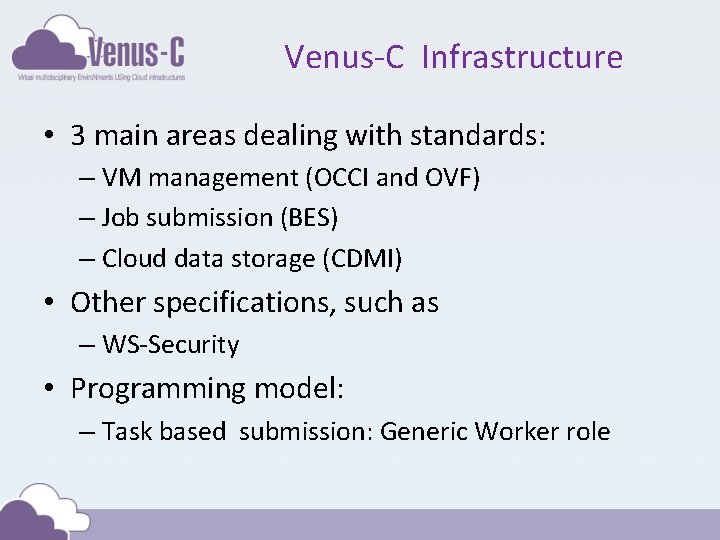
Venus-C Infrastructure • 3 main areas dealing with standards: – VM management (OCCI and OVF) – Job submission (BES) – Cloud data storage (CDMI) • Other specifications, such as – WS-Security • Programming model: – Task based submission: Generic Worker role

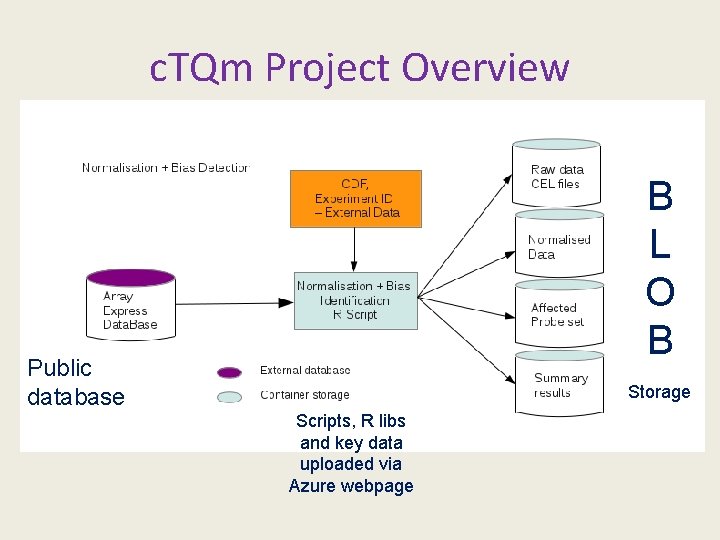
c. TQm Project Overview B L O B Public database Storage Scripts, R libs and key data uploaded via Azure webpage

Cloud / Grid Interfaces Amazon EC 2: Command line interface into Linux terminal NGS: Portal or Command Line to Linux machine Azure: Webpage interface to a Windows machine, Visual Studio 2010, C#


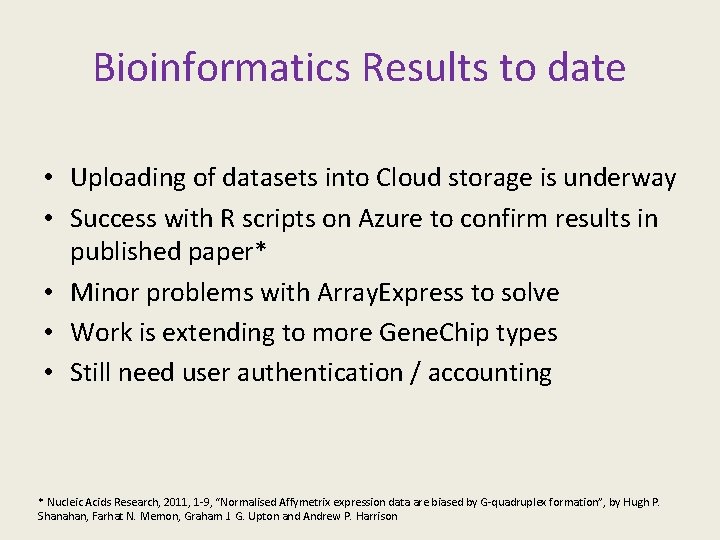
Bioinformatics Results to date • Uploading of datasets into Cloud storage is underway • Success with R scripts on Azure to confirm results in published paper* • Minor problems with Array. Express to solve • Work is extending to more Gene. Chip types • Still need user authentication / accounting * Nucleic Acids Research, 2011, 1 -9, “Normalised Affymetrix expression data are biased by G-quadruplex formation”, by Hugh P. Shanahan, Farhat N. Memon, Graham J. G. Upton and Andrew P. Harrison

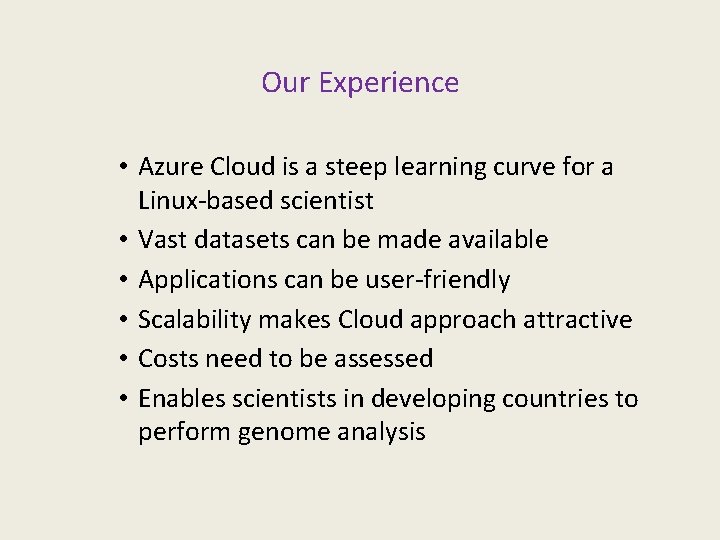
Our Experience • Azure Cloud is a steep learning curve for a Linux-based scientist • Vast datasets can be made available • Applications can be user-friendly • Scalability makes Cloud approach attractive • Costs need to be assessed • Enables scientists in developing countries to perform genome analysis

Analysis of Affymetrix expression data using R on Azure Cloud Acknowledgements and thanks to: Dr Andrew Harrison, University of Essex Dr Hugh Shanahan, Royal Holloway, University of London Department of Mathematical Sciences, University of Essex European Commission’s 7 th Framework Programme Microsoft and Venus-C project Organisers