Differential Expression Analysis Using RNASeq Diff Expression Analysis


- Slides: 11

Differential Expression Analysis Using RNA-Seq

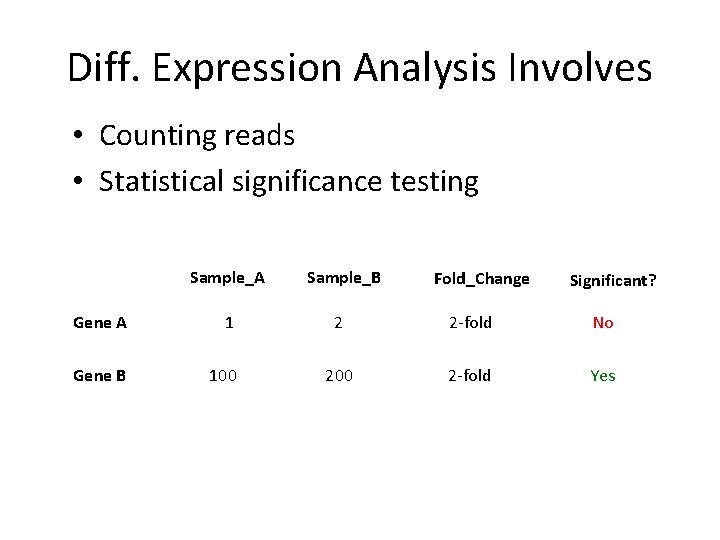
Diff. Expression Analysis Involves • Counting reads • Statistical significance testing Sample_A Gene A 1 Gene B 100 Sample_B Fold_Change Significant? 2 2 -fold No 200 2 -fold Yes

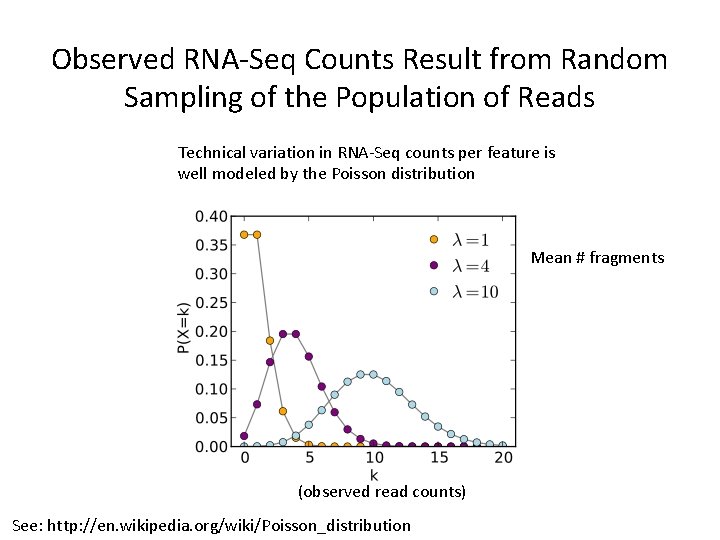
Observed RNA-Seq Counts Result from Random Sampling of the Population of Reads Technical variation in RNA-Seq counts per feature is well modeled by the Poisson distribution Mean # fragments (observed read counts) See: http: //en. wikipedia. org/wiki/Poisson_distribution

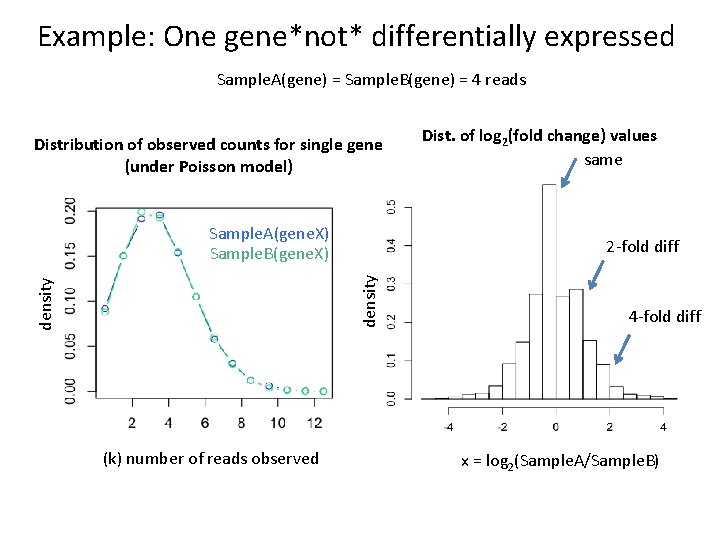
Example: One gene*not* differentially expressed Sample. A(gene) = Sample. B(gene) = 4 reads Distribution of observed counts for single gene (under Poisson model) Sample. A(gene. X) Sample. B(gene. X) 2 -fold diff density (k) number of reads observed Dist. of log 2(fold change) values same 4 -fold diff x = log 2(Sample. A/Sample. B)

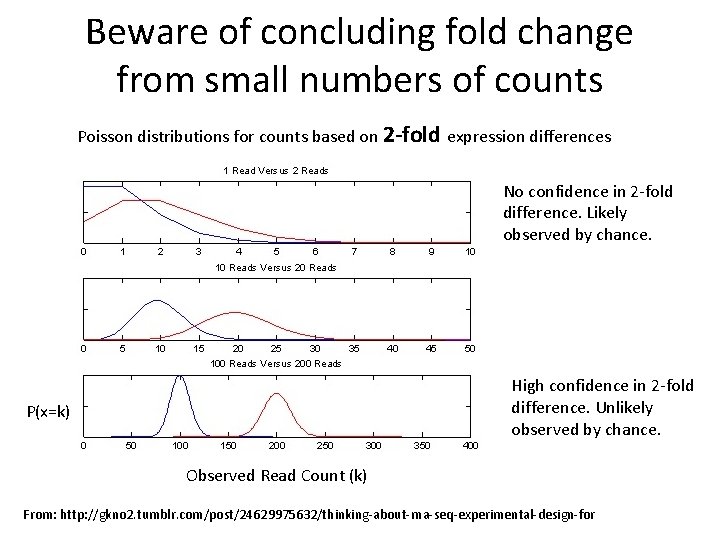
Beware of concluding fold change from small numbers of counts Poisson distributions for counts based on 2 -fold expression differences No confidence in 2 -fold difference. Likely observed by chance. High confidence in 2 -fold difference. Unlikely observed by chance. P(x=k) Observed Read Count (k) From: http: //gkno 2. tumblr. com/post/24629975632/thinking-about-rna-seq-experimental-design-for

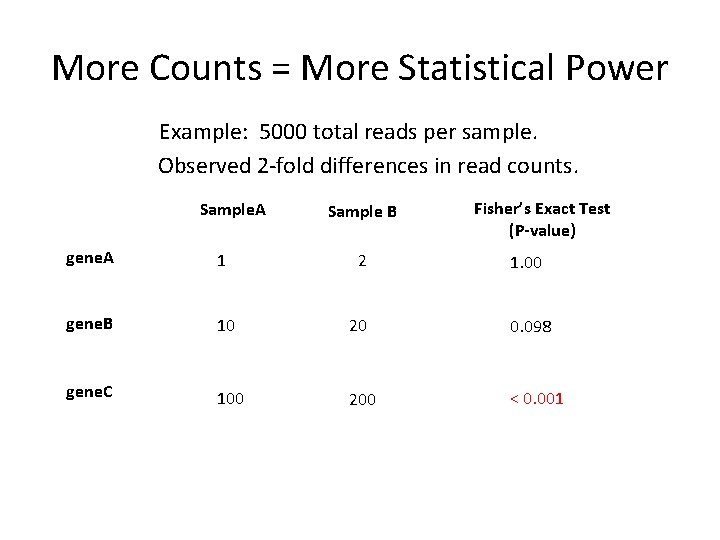
More Counts = More Statistical Power Example: 5000 total reads per sample. Observed 2 -fold differences in read counts. Sample. A Sample B Fisher’s Exact Test (P-value) gene. A 1 2 1. 00 gene. B 10 20 0. 098 gene. C 100 200 < 0. 001

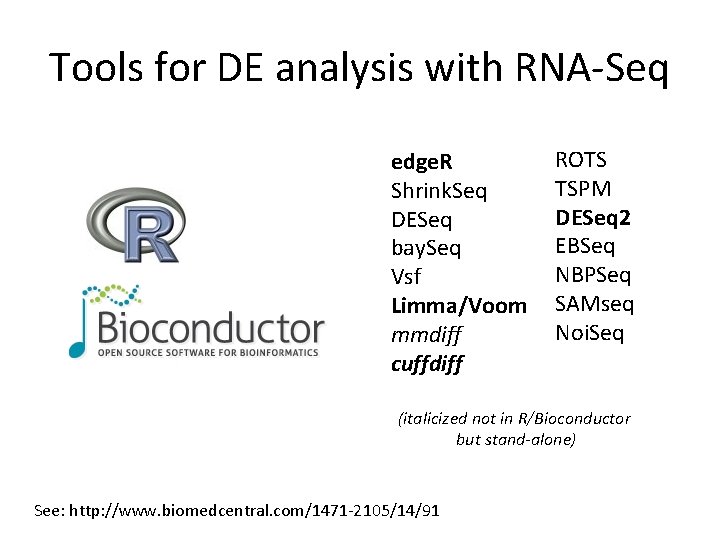
Tools for DE analysis with RNA-Seq edge. R Shrink. Seq DESeq bay. Seq Vsf Limma/Voom mmdiff cuffdiff ROTS TSPM DESeq 2 EBSeq NBPSeq SAMseq Noi. Seq (italicized not in R/Bioconductor but stand-alone) See: http: //www. biomedcentral. com/1471 -2105/14/91

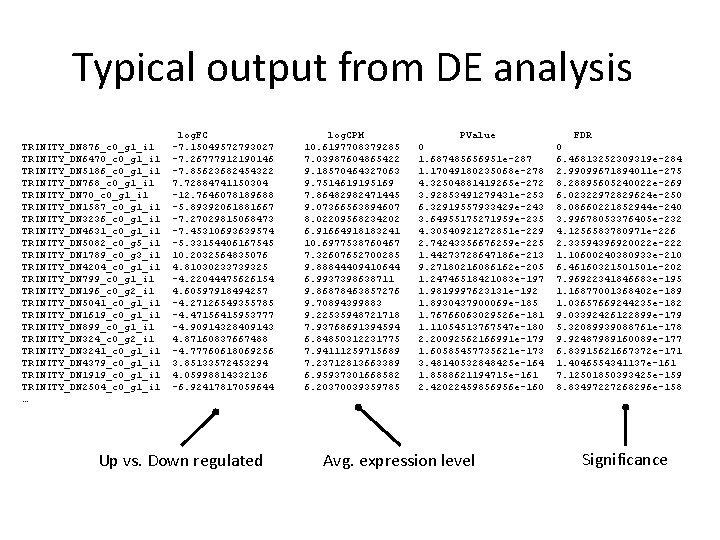
Typical output from DE analysis TRINITY_DN 876_c 0_g 1_i 1 TRINITY_DN 6470_c 0_g 1_i 1 TRINITY_DN 5186_c 0_g 1_i 1 TRINITY_DN 768_c 0_g 1_i 1 TRINITY_DN 70_c 0_g 1_i 1 TRINITY_DN 1587_c 0_g 1_i 1 TRINITY_DN 3236_c 0_g 1_i 1 TRINITY_DN 4631_c 0_g 1_i 1 TRINITY_DN 5082_c 0_g 5_i 1 TRINITY_DN 1789_c 0_g 3_i 1 TRINITY_DN 4204_c 0_g 1_i 1 TRINITY_DN 799_c 0_g 1_i 1 TRINITY_DN 196_c 0_g 2_i 1 TRINITY_DN 5041_c 0_g 1_i 1 TRINITY_DN 1619_c 0_g 1_i 1 TRINITY_DN 899_c 0_g 1_i 1 TRINITY_DN 324_c 0_g 2_i 1 TRINITY_DN 3241_c 0_g 1_i 1 TRINITY_DN 4379_c 0_g 1_i 1 TRINITY_DN 1919_c 0_g 1_i 1 TRINITY_DN 2504_c 0_g 1_i 1 … log. FC -7. 15049572793027 -7. 26777912190146 -7. 85623682454322 7. 72884741150304 -12. 7646078189688 -5. 89392061881667 -7. 27029815068473 -7. 45310693639574 -5. 33154406167545 10. 2032564835076 4. 81030233739325 -4. 22044475626154 4. 60597918494257 -4. 27126549355785 -4. 47156415953777 -4. 90914328409143 4. 87160837667488 -4. 77760618069256 3. 85133572453294 4. 05998814332136 -6. 92417817059644 Up vs. Down regulated log. CPM 10. 6197708379285 7. 03987604865422 9. 18570464327063 9. 7514619195169 7. 86482982471445 9. 07366563894607 8. 02209568234202 6. 91664918183241 10. 6977538760467 7. 32607652700285 9. 88844409410644 6. 9937398638711 9. 86878463857276 9. 70894399883 9. 22535948721718 7. 93768691394594 6. 84850312231775 7. 94111259715689 7. 23712813663389 6. 95937301668582 6. 20370039359785 PValue 0 1. 687485656951 e-287 1. 17049180235068 e-278 4. 32504881419265 e-272 3. 92853491279431 e-253 6. 32919557933429 e-243 3. 64955175271959 e-235 4. 30540921272851 e-229 2. 74243356676259 e-225 1. 44273728647186 e-213 9. 27180216086162 e-205 1. 24746518421083 e-197 1. 9819997623131 e-192 1. 8930437900069 e-185 1. 76766063029526 e-181 1. 11054513767547 e-180 2. 20092562166991 e-179 1. 60585457735621 e-173 3. 48140532848425 e-164 1. 8588621194715 e-161 2. 42022459856956 e-160 Avg. expression level FDR 0 6. 46813252309319 e-284 2. 99099671894011 e-275 8. 28895605240022 e-269 6. 02322972829624 e-250 8. 08660221852944 e-240 3. 99678053376405 e-232 4. 1256583780971 e-226 2. 33594396920022 e-222 1. 10600240380933 e-210 6. 46160321501501 e-202 7. 96922341846683 e-195 1. 16877001368402 e-189 1. 03657669244235 e-182 9. 03392426122899 e-179 5. 32089939088761 e-178 9. 92487989160089 e-177 6. 83915621667372 e-171 1. 4046554341137 e-161 7. 12501850393425 e-159 8. 83497227268296 e-158 Significance

Visualization of DE results and Expression Profiling

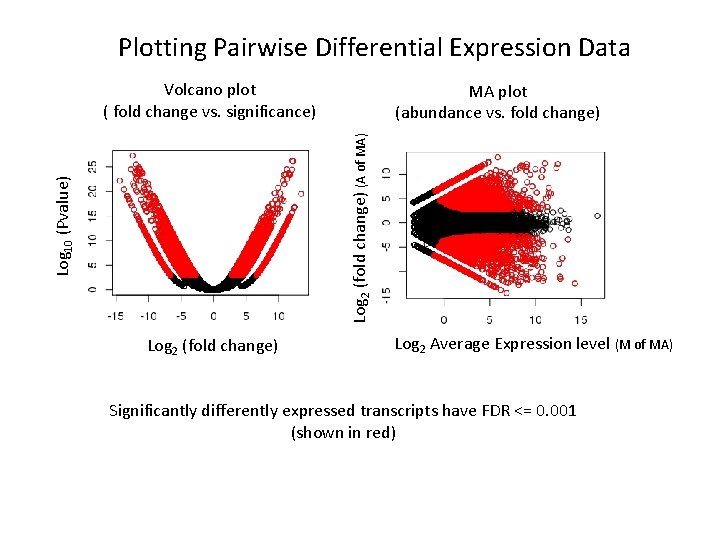
Plotting Pairwise Differential Expression Data Volcano plot ( fold change vs. significance) Log 10 (Pvalue) Log 2 (fold change) (A of MA) MA plot (abundance vs. fold change) Log 2 (fold change) Log 2 Average Expression level (M of MA) Significantly differently expressed transcripts have FDR <= 0. 001 (shown in red)

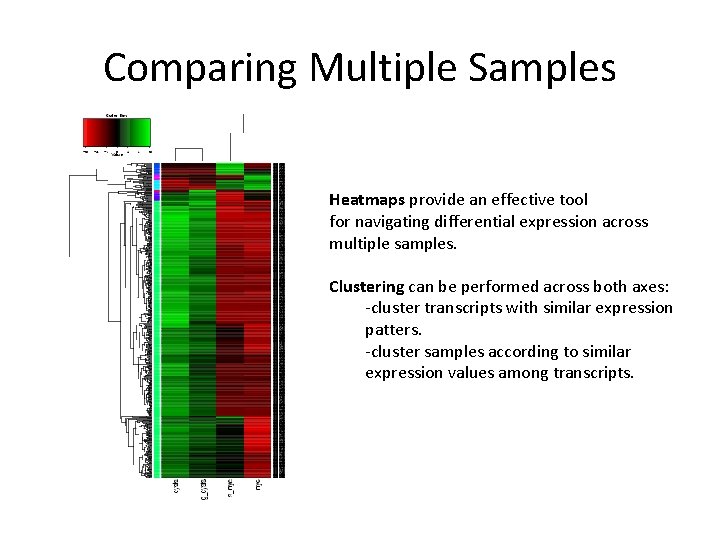
Comparing Multiple Samples Heatmaps provide an effective tool for navigating differential expression across multiple samples. Clustering can be performed across both axes: -cluster transcripts with similar expression patters. -cluster samples according to similar expression values among transcripts.