Affymetrix Gene Chip Data Analysis Chip concepts and





- Slides: 9

Affymetrix Gene. Chip Data Analysis • • Chip concepts and array design Improving intensity estimation from probe pairs level Clustering Motif discovering and preliminary result

How is Affymetrix Gene Chip made?

Affymetrix Gene Chip Procedure • Target preparation fragmented Isolate m. RNA c. DNA c. RNA 35~200 base fragments (12 -15 ug) biotinylated • Target Hybridization: 200 ml cocktail probe array chip hybridization (16 hours at 40~60°C) • Washing and Staining • Probe array scan, Data analysis, and Interpretation

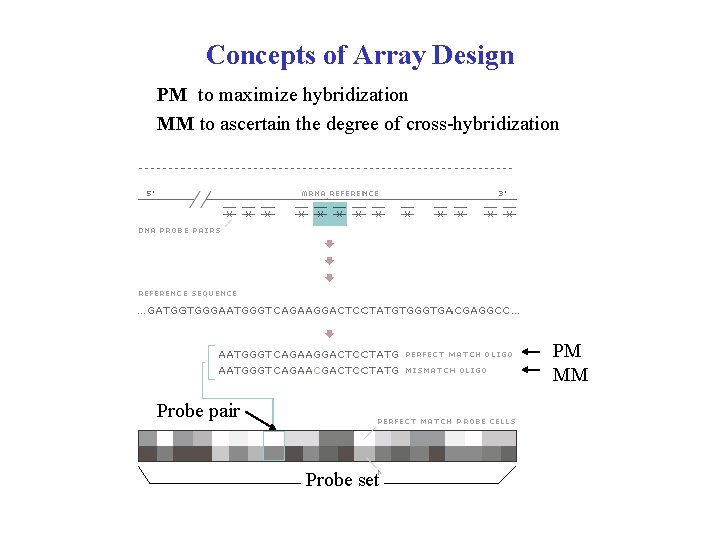
Concepts of Array Design PM to maximize hybridization MM to ascertain the degree of cross-hybridization PM MM Probe pair Probe set

Signal Intensity Estimation • Absolute analysis average difference (PM-MM) / pairs used • Comparison analysis experimental subtract baseline average difference • Probe pairs variation probe pairs outlier (cross-hybridization) arrays outlier (image contamination) • Probe pairs level adjustment Wing Wong Lab d. Chip: least square fitting model built from multiple chips Affymetrix MAS 5. 0: imputation

Clustering • Affymetrix DMT examine the SOM, are the clusters tight ? experiment primarily with "Number of Epochs", "Alpha_i" and "Sigma_i" parameters to reduce error. multiple level clustering examine filtering parameters to adjust min and max value. (default is 20 and 20000) • Wing H. Wong Lab d. Chip from image file to outlier detection and clustering • MIT Whitehead Institute Gene. Cluster • Stanford Cluster and Treeview two way clustering (genes and/or arrays) find profile pattern not found in SOM clustering

Motif discovering • Motif discovery and matching tools Basic assumption: a cluster of co-regulated genes is regulated by the same transcription factors and the genes of a given cluster share common regulatory motifs Gibbs Motif Sampler Neuwald et al. 1995 Align. ACE based on Gibbs Sampling, optimized for finding multiple motifs and for both strands RSA-tools motif finding among non-coding regions in cress, yeast, fly, worm Plant. CARE search known motifs among 417 different plant transcription sites TFSEARCH search known motifs in vertebrate, arthropod, plant, yeast

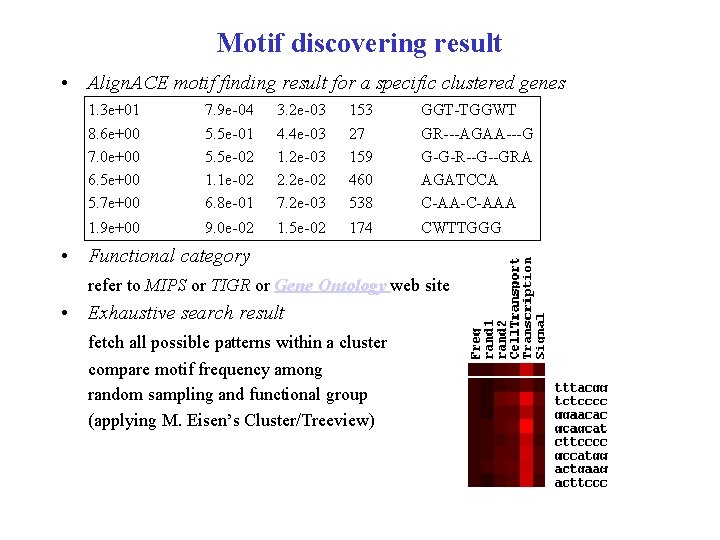
Motif discovering result • Align. ACE motif finding result for a specific clustered genes 1. 3 e+01 8. 6 e+00 7. 0 e+00 6. 5 e+00 5. 7 e+00 7. 9 e-04 5. 5 e-01 5. 5 e-02 1. 1 e-02 6. 8 e-01 3. 2 e-03 4. 4 e-03 1. 2 e-03 2. 2 e-02 7. 2 e-03 153 27 159 460 538 GGT-TGGWT GR---AGAA---G G-G-R--G--GRA AGATCCA C-AA-C-AAA 1. 9 e+00 9. 0 e-02 1. 5 e-02 174 CWTTGGG • Functional category refer to MIPS or TIGR or Gene Ontology web site • Exhaustive search result fetch all possible patterns within a cluster compare motif frequency among random sampling and functional group (applying M. Eisen’s Cluster/Treeview)

Supportive Information • Schroeder lab (UCSD) sequence retrieval and direct MEME motif finding • Gene Microarray Pathway Profiler genechip array statistical consideration • MIT CCR HHMI Biopolymers Laboratory • Databases and software tools for gene expression software, databases, mail lists