Tumour Modifier Genes 1 STRATEGY KISS OF DEATH




- Slides: 27

Tumour Modifier Genes 1. STRATEGY (KISS OF DEATH) 2. SOME RESULTS 3. WHY DO HUMAN ASSOCIATION STUDIES NOT WORK (WELL)?

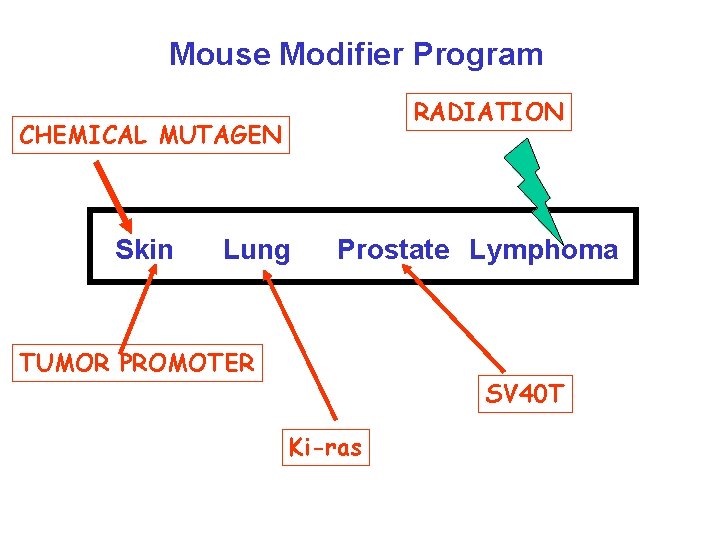
Mouse Modifier Program RADIATION CHEMICAL MUTAGEN Skin Lung Prostate Lymphoma TUMOR PROMOTER SV 40 T Ki-ras

GENETIC APPROACHES TO TUMOR MODIFIERS 1. INTRASPECIFIC MUSCULUS X MUSCULUS CROSSES backcross intercross recombinant inbred recombinant congenic 2. INTERSPECIFIC SPRETUS X MUSCULUS CROSSES 3. ADVANCED MULTISTRAIN INTERCROSS 4 OUTBRED POPULATIONS

INTERSPECIFIC SPRETUS-MUSCULUS CROSS ADVANTAGES 1. GENETIC DIVERGENCE - extreme phenotypic differences 2. DOMINANT RESISTANCE GENES – reduced complexity 3 HIGHLY POLYMORPHIC- DISADVANTAGES 1. Male F 1 s are sterile 2. Can be difficult to breed mapping is easy

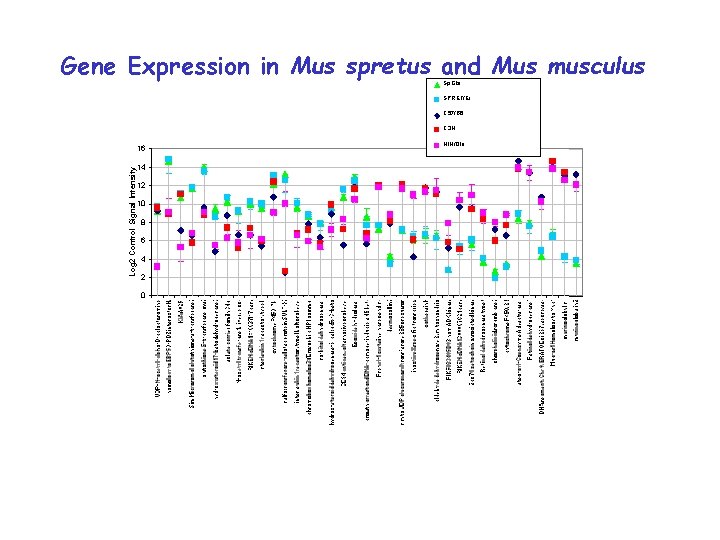
Gene Expression in Mus spretus and Mus musculus Sp. Gla SPRET/Ei C 57/B 6 C 3 H 16 Log 2 Control Signal Intensity 14 12 10 8 6 4 2 0 NIH/Ola

Dominant resistance genes in mus spretus Mus spretus are resistant to tumor development in the SKIN LUNG COLON LIVER LYMPHOID SYSTEM

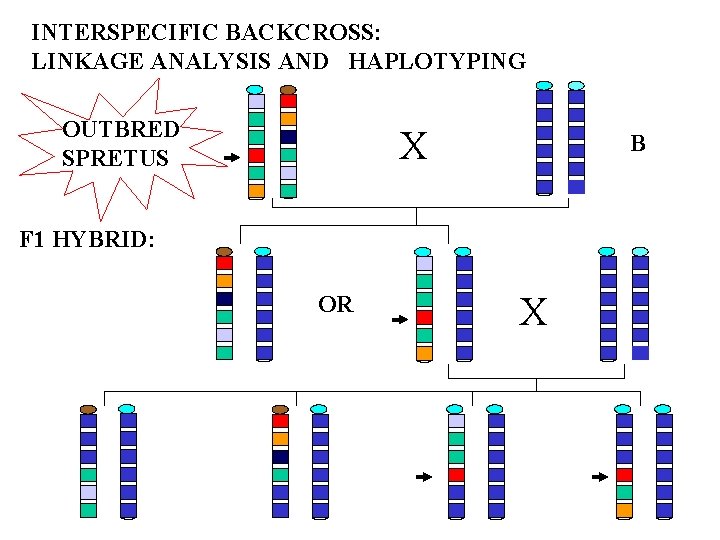
INTERSPECIFIC BACKCROSS: LINKAGE ANALYSIS AND HAPLOTYPING OUTBRED SPRETUS X B F 1 HYBRID: OR X

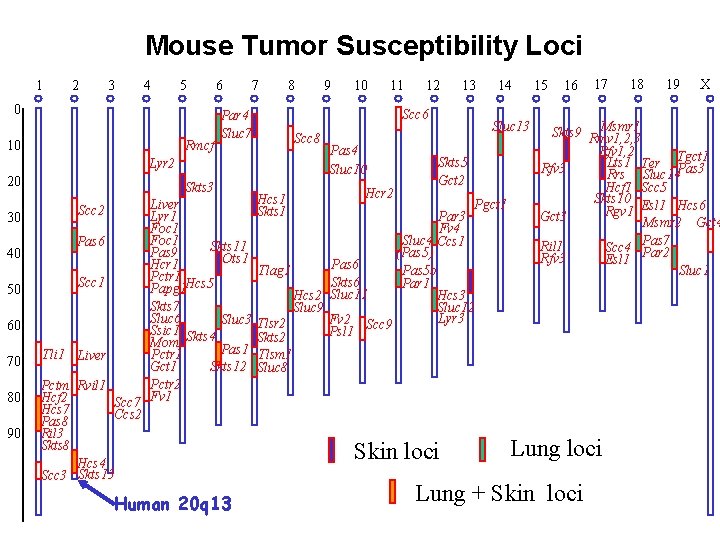
Mouse Tumor Susceptibility Loci 1 2 3 4 5 0 10 Rmcf 6 7 30 40 50 Skts 3 Scc 2 Pas 6 Scc 1 60 70 80 90 Tli 1 Liver Lyr 1 Foc 1 Skts 11 Pas 9 Ots 1 Hcr 1 Pctr 1 Hcs 5 Papg 1 Skts 7 Sluc 6 Sluc 3 Ssic 1 Skts 4 Mom 1 Pas 1 Pctr 1 Gct 1 Skts 12 Pctr 2 Scc 7 Fv 1 Ccs 2 Pctm Rvil 1 Hcf 2 Hcs 7 Pas 8 Ril 3 Skts 8 Hcs 4 Scc 3 Skts 13 Human 20 q 13 9 10 11 12 13 Scc 6 Par 4 Sluc 7 Scc 8 Lyr 2 20 8 Hcs 1 Skts 1 Pas 4 Sluc 10 Hcr 2 14 Sluc 13 Skts 5 Gct 2 Pgct 1 Par 3 Fv 4 Sluc 4 Ccs 1 (Pas 5) Pas 6 Tlag 1 Pas 5 b Skts 6 Par 1 Hcs 2 Sluc 11 Hcs 3 Sluc 12 Sluc 9 Lyr 3 Fv 2 Scc 9 Tlsr 2 Psl 1 Skts 2 Tlsm 1 Sluc 8 Skin loci 15 16 17 18 19 X Msmr 1 Skts 9 Rmv 1, 2, 3 Rfv 1, 2 Lts 1 Ter Tgct 1 Rfv 3 Rrs Sluc 14 Pas 3 Hcf 1 Scc 5 Skts 10 Esl 1 Hcs 6 Rgv 1 Gct 3 Msmr 2 Gct 4 Ril 1 Scc 4 Pas 7 Rfv 3 Esl 1 Par 2 Sluc 1 Lung loci Lung + Skin loci

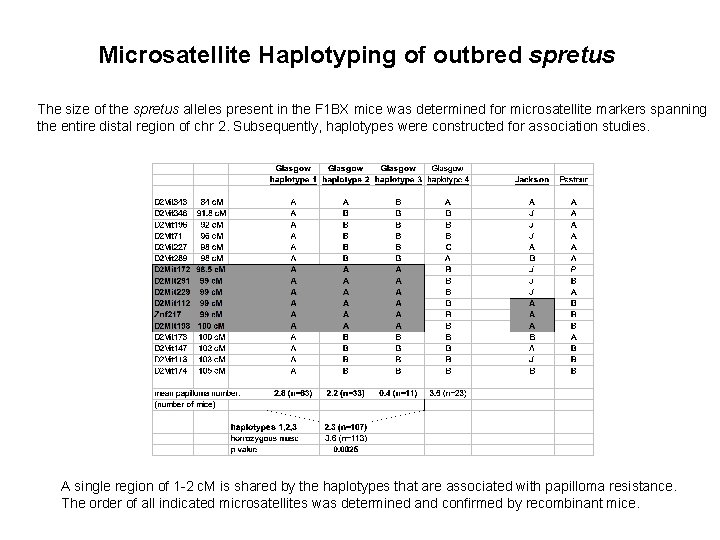
Microsatellite Haplotyping of outbred spretus The size of the spretus alleles present in the F 1 BX mice was determined for microsatellite markers spanning the entire distal region of chr 2. Subsequently, haplotypes were constructed for association studies. A single region of 1 -2 c. M is shared by the haplotypes that are associated with papilloma resistance. The order of all indicated microsatellites was determined and confirmed by recombinant mice.

mouse human

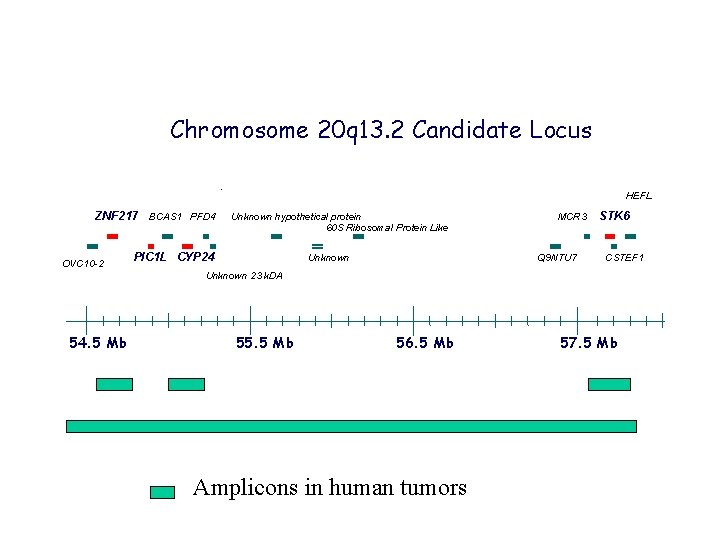
Chromosome 20 q 13. 2 Candidate Locus HEFL ZNF 217 BCAS 1 PFD 4 OVC 10 -2 Unknown hypothetical protein 60 S Ribosomal Protein Like PIC 1 L CYP 24 MCR 3 Q 9 NTU 7 Unknown STK 6 CSTEF 1 Unknown 23 k. DA 54. 5 Mb 55. 5 Mb 56. 5 Mb Amplicons in human tumors 57. 5 Mb

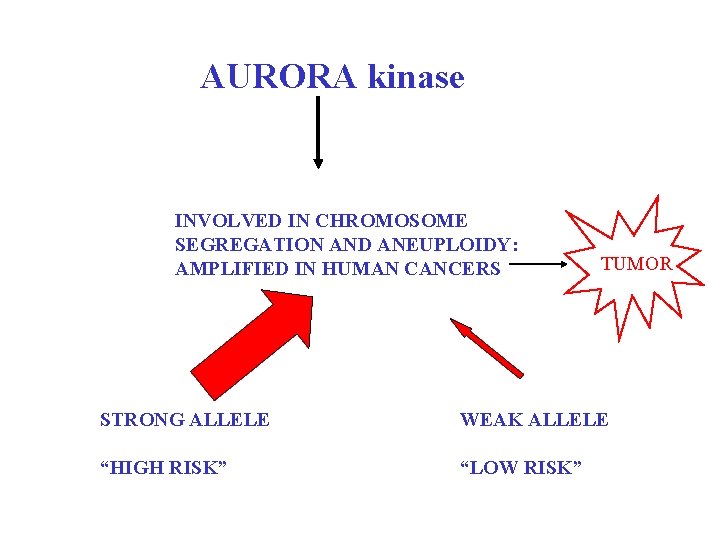
AURORA kinase INVOLVED IN CHROMOSOME SEGREGATION AND ANEUPLOIDY: AMPLIFIED IN HUMAN CANCERS TUMOR STRONG ALLELE WEAK ALLELE “HIGH RISK” “LOW RISK”

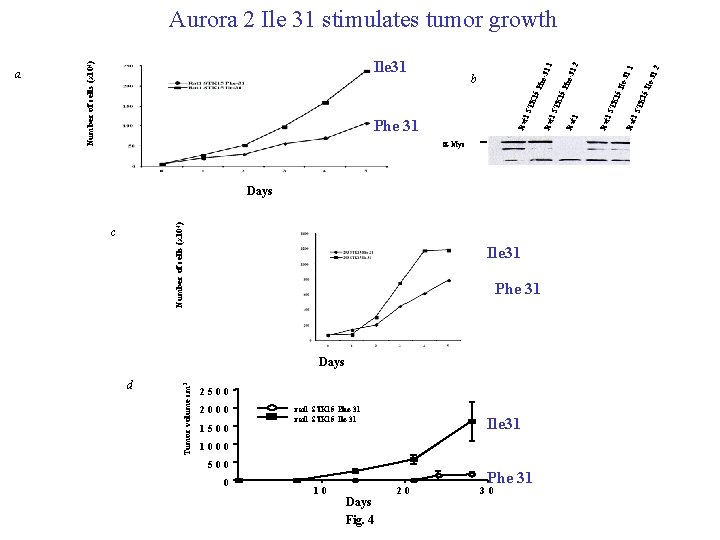
a-Myc Number of cells (x 104) Days c Ile 31 Phe 31 d Tumor volume cm 3 Days 2500 2000 1500 rat 1 STK 15 Phe-31 rat 1 STK 15 Ile-31 Ile 31 1000 500 0 10 Days Fig. 4 20 Phe 31 30 2 15 15 TK Rat 1 S STK Rat 1 Ile- Ile 3 31. 1. 1 -31. 15 STK Rat 1 Phe 31 Rat 1 STK 15 Phe b Phe -31. 1 Ile 31 Number of cells (x 104) a 2 Aurora 2 Ile 31 stimulates tumor growth

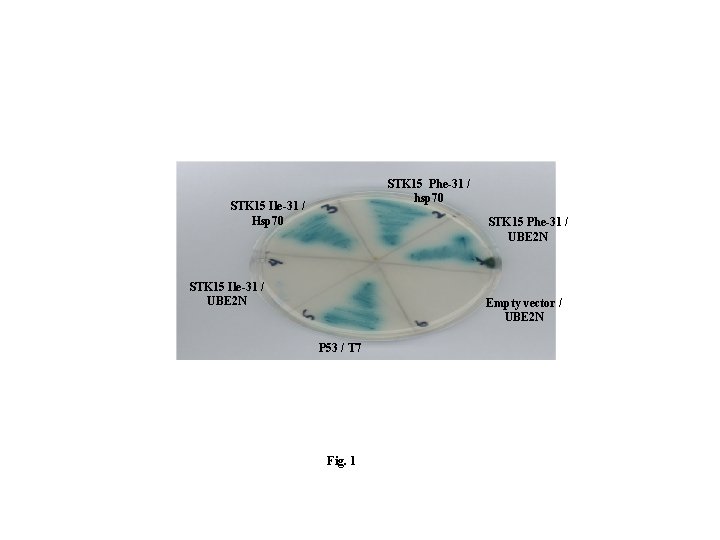
STK 15 Phe-31 / hsp 70 STK 15 Ile-31 / Hsp 70 STK 15 Phe-31 / UBE 2 N STK 15 Ile-31 / UBE 2 N Empty vector / UBE 2 N P 53 / T 7 Fig. 1

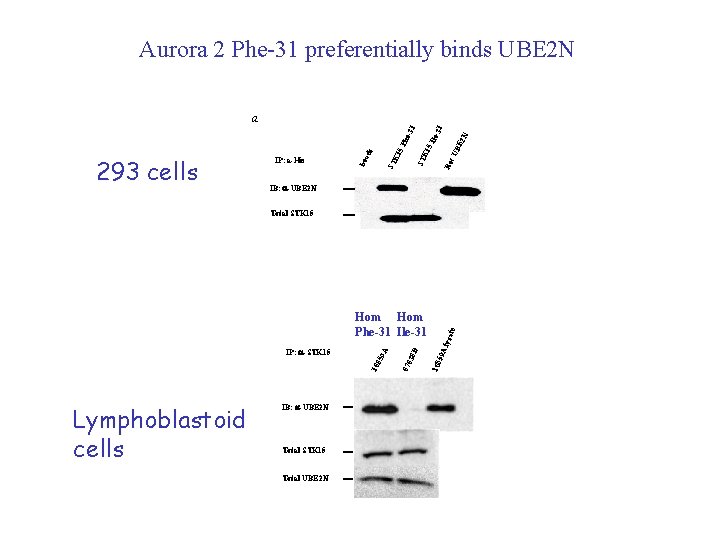
Aurora 2 Phe-31 preferentially binds UBE 2 N 1 BE 2 N le-3 Re c. U 5 I ST K 1 5 P ST K 1 bea 293 cells IP: a-His ds he- 31 a IB: a-UBE 2 N Lymphoblastoid cells IB: a-UBE 2 N Total STK 15 Total UBE 2 N lysa 59 A 108 070 38 D IP: a-STK 15 108 59 A Hom Phe-31 Ile-31 te Total STK 15

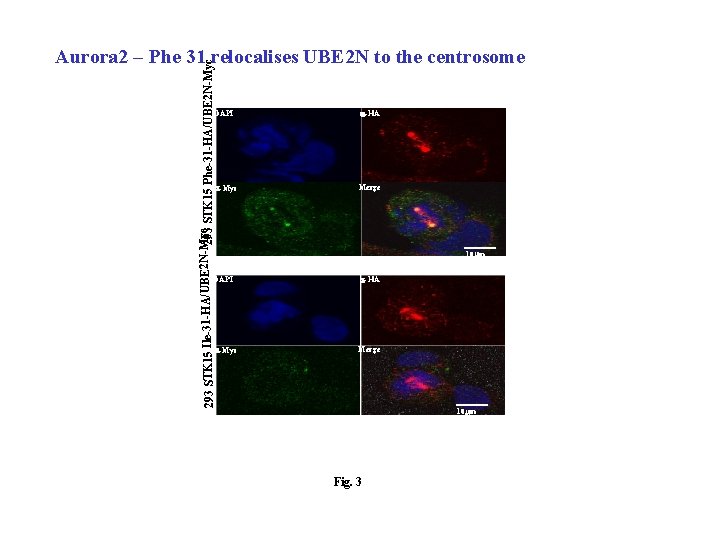
yc STK 15 Phe-31 -HA/UBE 2 N-Myc 293 STK 15 Ile-31 -HA/UBE 2 N-M Aurora 2 – Phe 31 relocalises UBE 2 N to the centrosome DAPI a-HA a-Myc Merge 10 mm Fig. 3

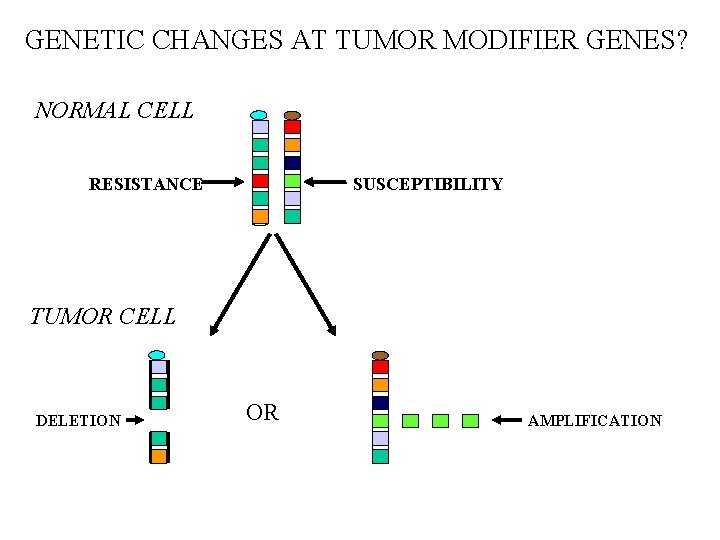
GENETIC CHANGES AT TUMOR MODIFIER GENES? NORMAL CELL RESISTANCE SUSCEPTIBILITY TUMOR CELL DELETION OR AMPLIFICATION

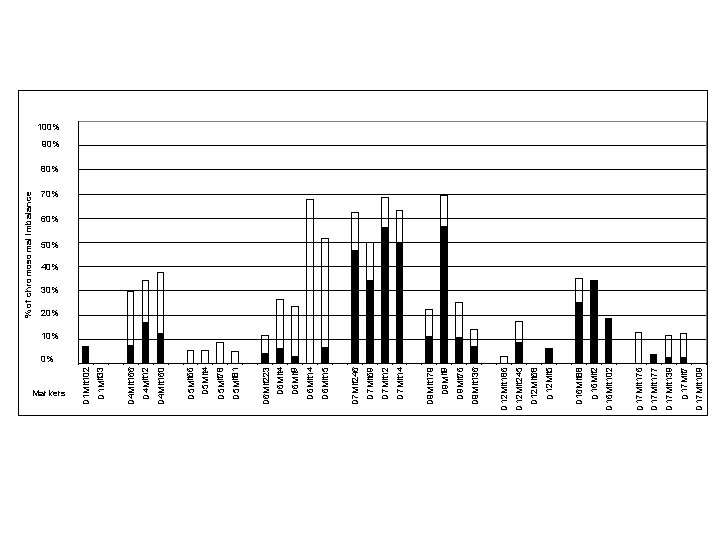
Markers D 17 Mit 109 D 17 Mit 7 D 17 Mit 139 D 17 Mit 177 D 17 Mit 176 D 16 Mit 102 D 16 Mit 88 D 12 Mit 5 D 12 Mit 68 D 12 Mit 245 D 12 Mit 186 D 9 Mit 136 D 9 Mit 76 D 9 Mit 9 D 9 Mit 179 D 7 Mit 14 D 7 Mit 12 D 7 Mit 69 D 7 Mit 246 D 6 Mit 15 D 6 Mit 14 D 6 Mit 9 D 6 Mit 4 D 6 Mit 223 D 5 Mit 81 D 5 Mit 78 D 5 Mit 4 D 5 Mit 66 D 4 Mit 160 D 4 Mit 12 D 4 Mit 166 D 1 Mit 33 D 1 Mit 102 % of chromosomal imbalance 100% 90% 80% 70% 60% 50% 40% 30% 20% 10% 0%

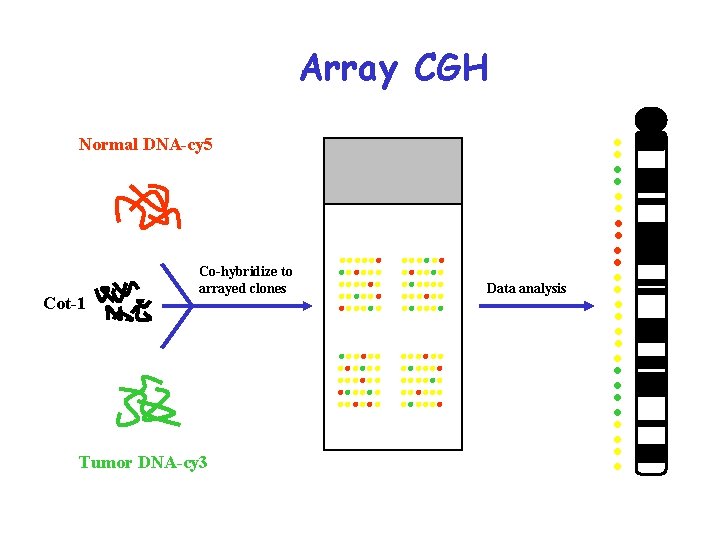
Array CGH Normal DNA-cy 5 Cot-1 Co-hybridize to arrayed clones Tumor DNA-cy 3 Data analysis

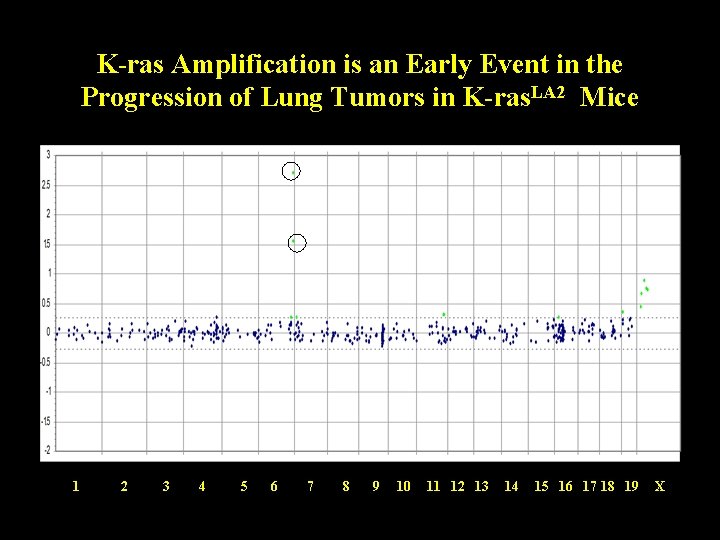
K-ras Amplification is an Early Event in the Progression of Lung Tumors in K-ras. LA 2 Mice 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 X

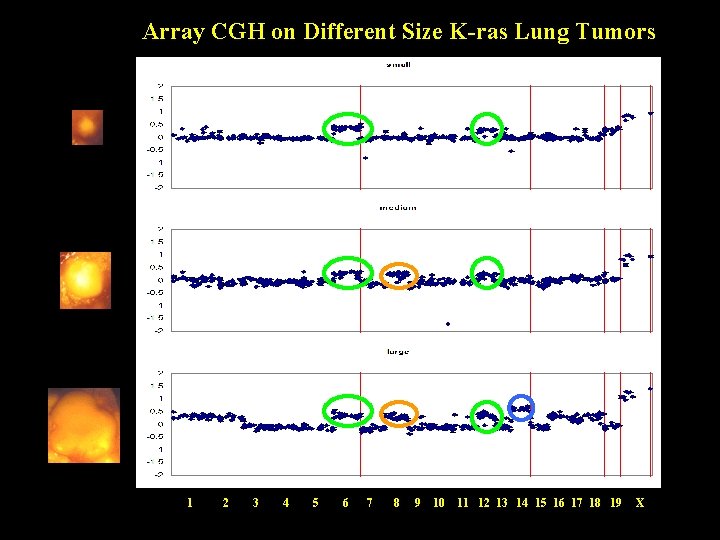
Array CGH on Different Size K-ras Lung Tumors 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 X

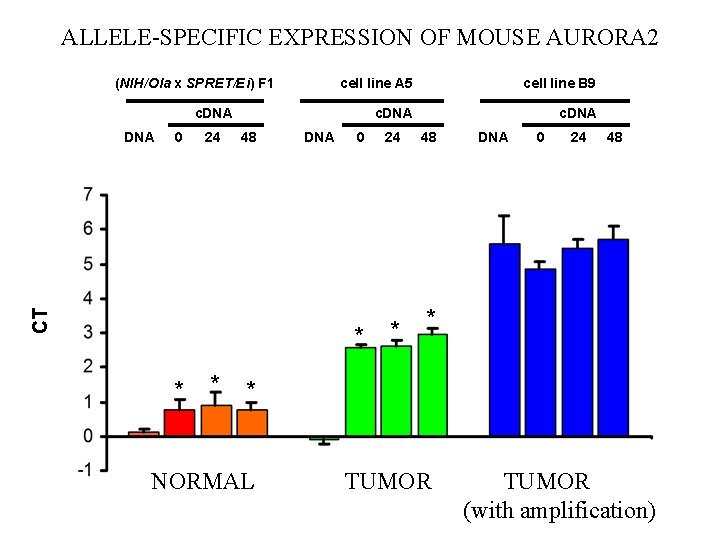
ALLELE-SPECIFIC EXPRESSION OF MOUSE AURORA 2 (NIH/Ola x SPRET/Ei) F 1 cell line A 5 cell line B 9 c. DNA 0 24 48 CT DNA * * DNA 0 24 * * 48 DNA 0 24 48 * * NORMAL TUMOR (with amplification)

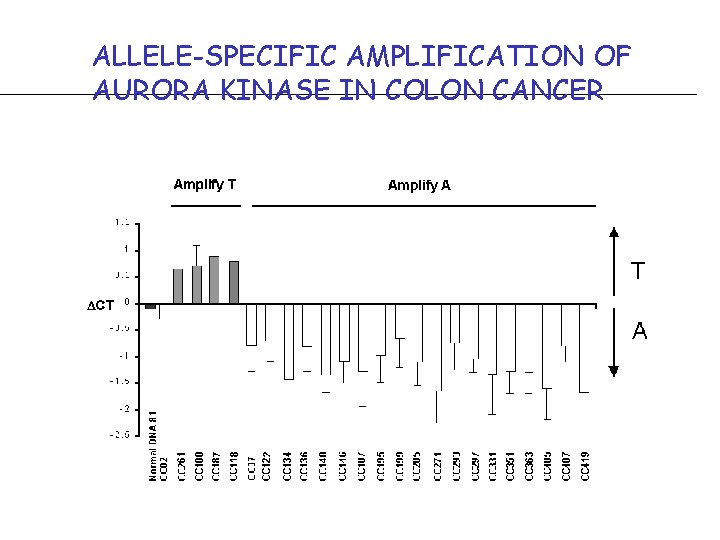
ALLELE-SPECIFIC AMPLIFICATION OF AURORA KINASE IN COLON CANCER Fig 5. Colon tumor amplification of Stk 6 alleles

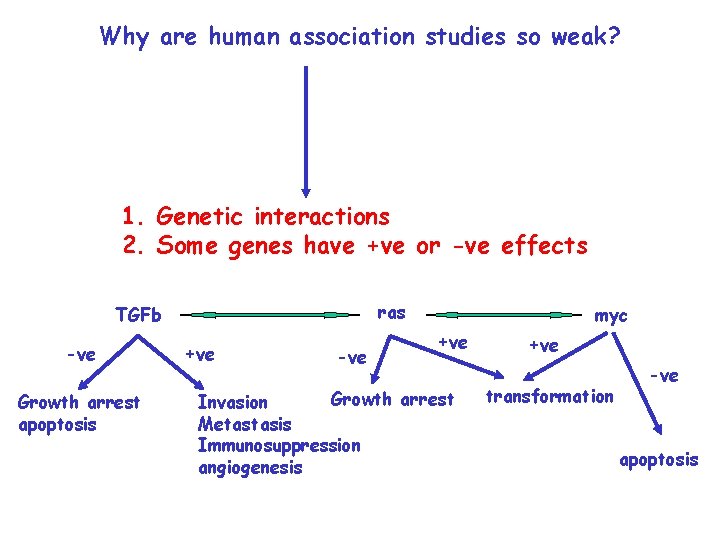
Why are human association studies so weak? 1. Genetic interactions 2. Some genes have +ve or -ve effects ras TGFb -ve Growth arrest apoptosis +ve -ve myc +ve Growth arrest Invasion Metastasis Immunosuppression angiogenesis +ve transformation -ve apoptosis

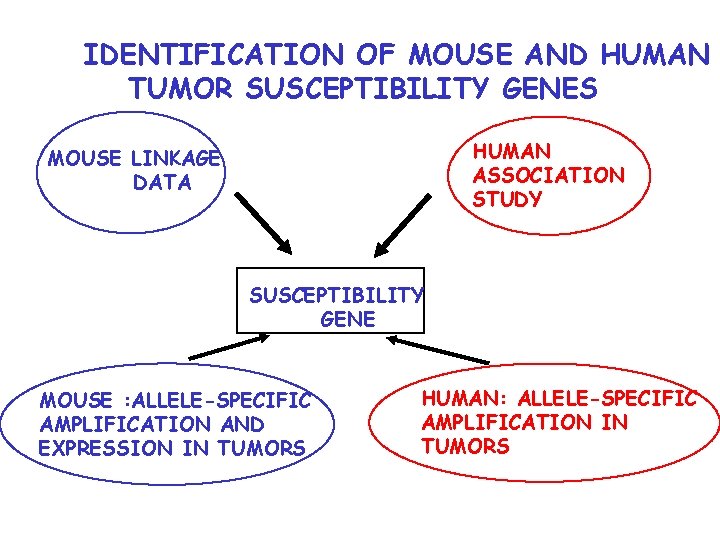
IDENTIFICATION OF MOUSE AND HUMAN TUMOR SUSCEPTIBILITY GENES HUMAN ASSOCIATION STUDY MOUSE LINKAGE DATA SUSCEPTIBILITY GENE MOUSE : ALLELE-SPECIFIC AMPLIFICATION AND EXPRESSION IN TUMORS HUMAN: ALLELE-SPECIFIC AMPLIFICATION IN TUMORS

ACKNOWLEDGEMENTS Mandy Toland Jian-Hua Mao Jin-wei Yuan Jeff Hsu Di Wu Reyno Delrosario John de Koning Hiroki Nagase Joe Gray Graeme Hodgson June Chan Jing Ma - Harvard Tyler Jacks – MIT Spiros Linardopoulos. London Ponder Lab-Cambridge NCI MOUSE MODELS OF HUMAN CANCER CONSORTIUM

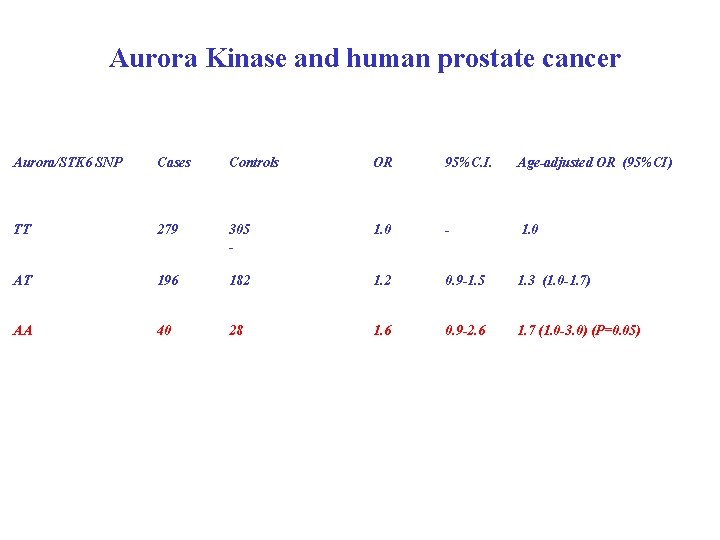
Aurora Kinase and human prostate cancer Aurora/STK 6 SNP Cases Controls OR 95%C. I. Age-adjusted OR (95%CI) TT 279 305 - 1. 0 AT 196 182 1. 2 0. 9 -1. 5 1. 3 (1. 0 -1. 7) AA 40 28 1. 6 0. 9 -2. 6 1. 7 (1. 0 -3. 0) (P=0. 05)
 Brain tumors
Brain tumors Giant cell tumor
Giant cell tumor Adenoma
Adenoma Polygenic inheritance
Polygenic inheritance Linked genes and unlinked genes
Linked genes and unlinked genes Glomerulus
Glomerulus Clamped wished ticked posed
Clamped wished ticked posed Difference between dangling and misplaced modifier
Difference between dangling and misplaced modifier Somatic and molecular death
Somatic and molecular death Transnational strategy vs global strategy
Transnational strategy vs global strategy Listening
Listening Corporate strategy and business strategy
Corporate strategy and business strategy Aligning hr strategy with business strategy
Aligning hr strategy with business strategy Directional strategies in strategic management
Directional strategies in strategic management Global matrix structure
Global matrix structure Process of crafting and executing strategy
Process of crafting and executing strategy Global strategy
Global strategy International or multinational
International or multinational Chase strategy examples
Chase strategy examples Strategy formulation vs strategy implementation
Strategy formulation vs strategy implementation Cdc genes
Cdc genes Penetrance and expressivity
Penetrance and expressivity Genes located on the sex chromosomes
Genes located on the sex chromosomes Dna and genes chapter 11
Dna and genes chapter 11 Non protein coding genes
Non protein coding genes Punnett square blood type ab and o
Punnett square blood type ab and o Características del codigo genetico
Características del codigo genetico Jumping genes
Jumping genes