Research Group Overview Joel L Kaar Associate Professor


- Slides: 7

Research Group Overview Joel L. Kaar, Associate Professor Department of Chemical & Biological Engineering, University of Colorado Boulder

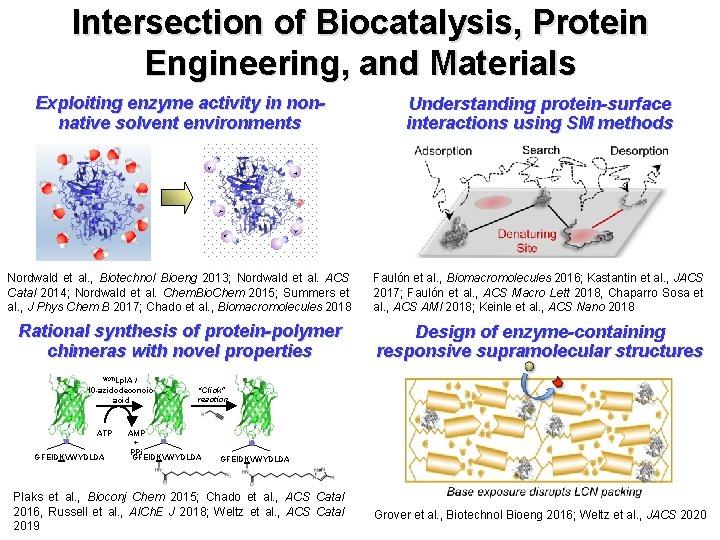
Intersection of Biocatalysis, Protein Engineering, and Materials Exploiting enzyme activity in nonnative solvent environments Understanding protein-surface interactions using SM methods Nordwald et al. , Biotechnol Bioeng 2013; Nordwald et al. ACS Catal 2014; Nordwald et al. Chem. Bio. Chem 2015; Summers et al. , J Phys Chem B 2017; Chado et al. , Biomacromolecules 2018 Faulón et al. , Biomacromolecules 2016; Kastantin et al. , JACS 2017; Faulón et al. , ACS Macro Lett 2018, Chaparro Sosa et al. , ACS AMI 2018; Keinle et al. , ACS Nano 2018 Rational synthesis of protein-polymer chimeras with novel properties Design of enzyme-containing responsive supramolecular structures W 37 ILpl. A / 10 -azidodeconoic acid ATP GFEIDKVWYDLDA “Click” reaction AMP + PPi GFEIDKVWYDLDA Plaks et al. , Bioconj Chem 2015; Chado et al. , ACS Catal 2016, Russell et al. , AICh. E J 2018; Weltz et al. , ACS Catal 2019 Grover et al. , Biotechnol Bioeng 2016; Weltz et al. , JACS 2020

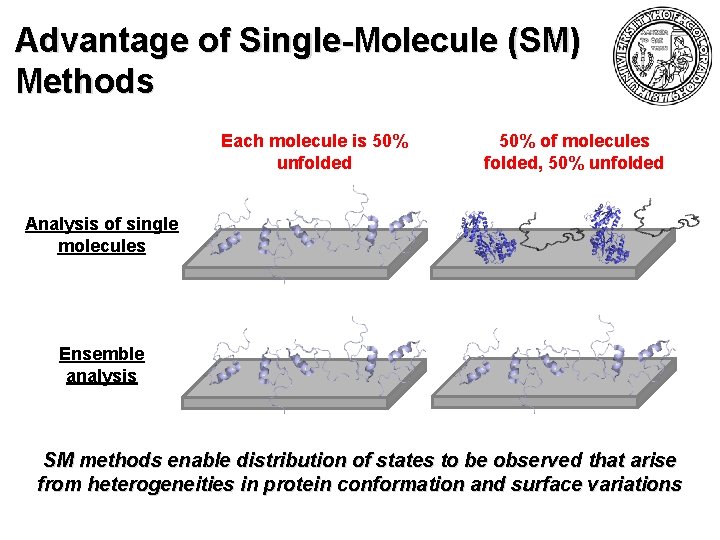
Advantage of Single-Molecule (SM) Methods Each molecule is 50% unfolded 50% of molecules folded, 50% unfolded Analysis of single molecules Ensemble analysis SM methods enable distribution of states to be observed that arise from heterogeneities in protein conformation and surface variations

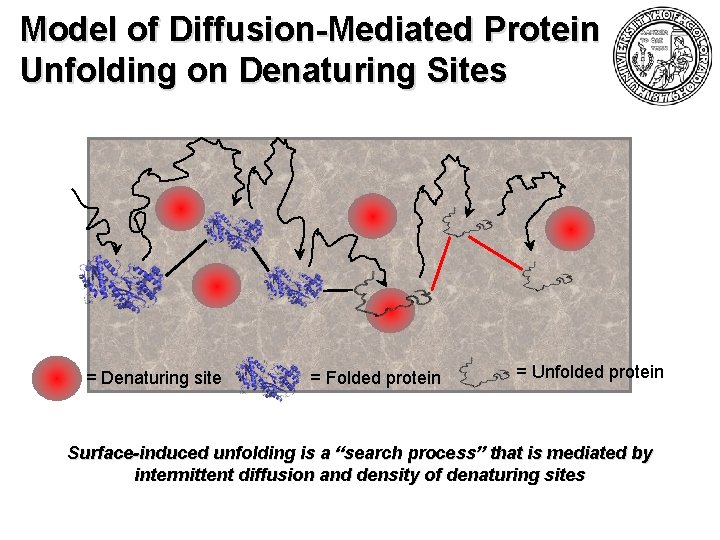
Model of Diffusion-Mediated Protein Unfolding on Denaturing Sites = Denaturing site = Folded protein = Unfolded protein Surface-induced unfolding is a “search process” that is mediated by intermittent diffusion and density of denaturing sites

Stabilizing FN by Copolymer Brushes Inhibits Macrophage Activation p = 0. 1 p = 0. 0001 p = 0. 003 p < 0. 0001 p = 0. 005 p = 0. 0002 p = 0. 6 Murine macrophages (RAW 264. 7) were cultured for 24 h on brushes after incubation in defined media containing FN PEG/PSB brushes significantly reduced the secretion of the pro-inflammatory cytokines IL-6 and TNF-a

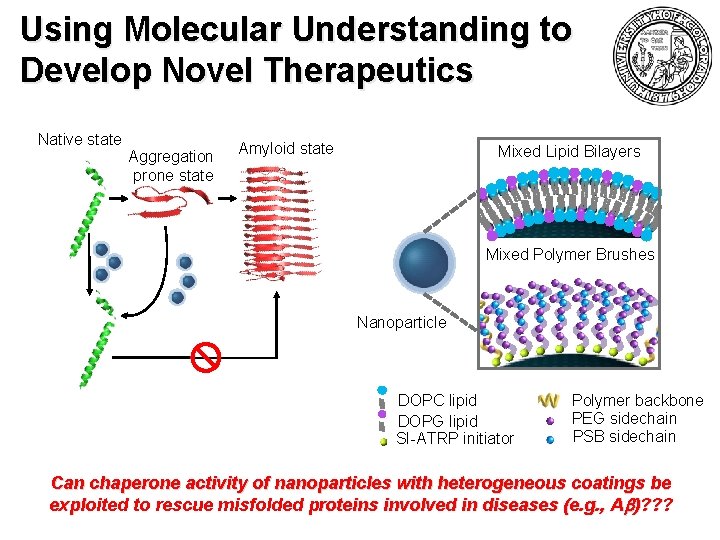
Using Molecular Understanding to Develop Novel Therapeutics Native state Aggregation prone state Amyloid state Mixed Lipid Bilayers Mixed Polymer Brushes Nanoparticle DOPC lipid DOPG lipid SI-ATRP initiator Polymer backbone PEG sidechain PSB sidechain Can chaperone activity of nanoparticles with heterogeneous coatings be exploited to rescue misfolded proteins involved in diseases (e. g. , A b)? ? ?

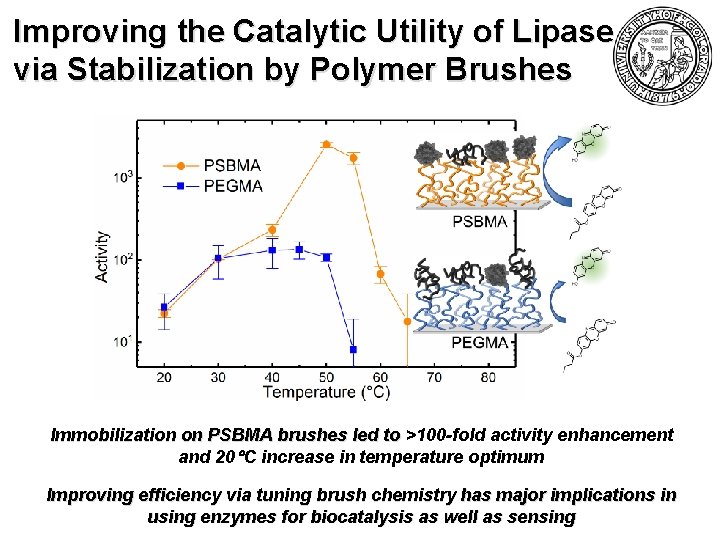
Improving the Catalytic Utility of Lipase via Stabilization by Polymer Brushes Immobilization on PSBMA brushes led to >100 -fold activity enhancement and 20 C increase in temperature optimum Improving efficiency via tuning brush chemistry has major implications in using enzymes for biocatalysis as well as sensing