Plasmid DNA Isolation Experiment Goals Extraction of plasmid




- Slides: 19

Plasmid DNA Isolation

Experiment Goals • Extraction of plasmid DNA from E. Coli. • Analyze plasmid DNA by agarose gel electrophoresis and spectrophotometer.

Introduction • Many types of bacteria contain plasmid DNA. • Plasmids are extrachromosomal, double-stranded circular DNA molecules separate from the chromosomal DNA. • Certain plasmids replicate independently of the chromosomal DNA and can be present in hundreds of copies per cell. • Generally containing 1, 000 to 100, 000 base pairs. • Even the largest plasmids are considerably smaller than the chromosomal DNA of the bacterium, which can contain several million base pairs.

Classification of plasmids by function There are five main classes • Fertility-F-plasmids, Facilitate bacterial conjugation • Resistance-R-plasmids, which contain genes that can build a resistance against antibiotics or poisons. • Col-plasmids, which contain genes that code for bacteriocins, proteins that can kill other bacteria. • Degradative plasmids, which enable the digestion of unusual substances, e. g. , toluene or salicylic acid. • Virulence plasmids, which turn the bacterium into a pathogen.

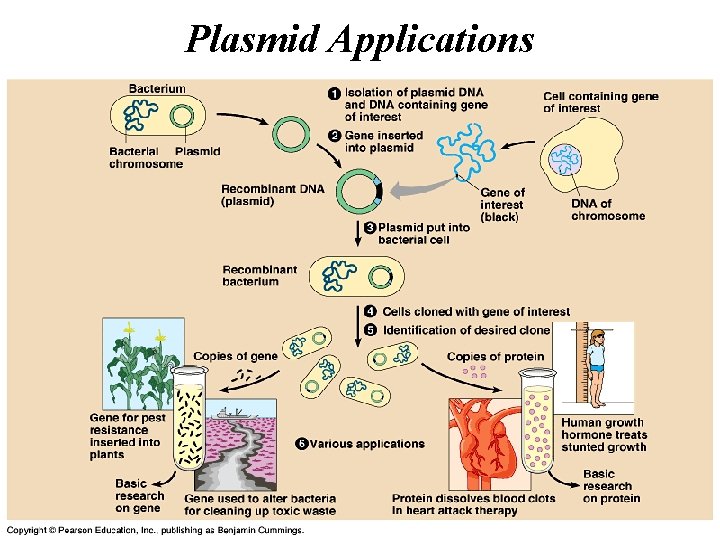
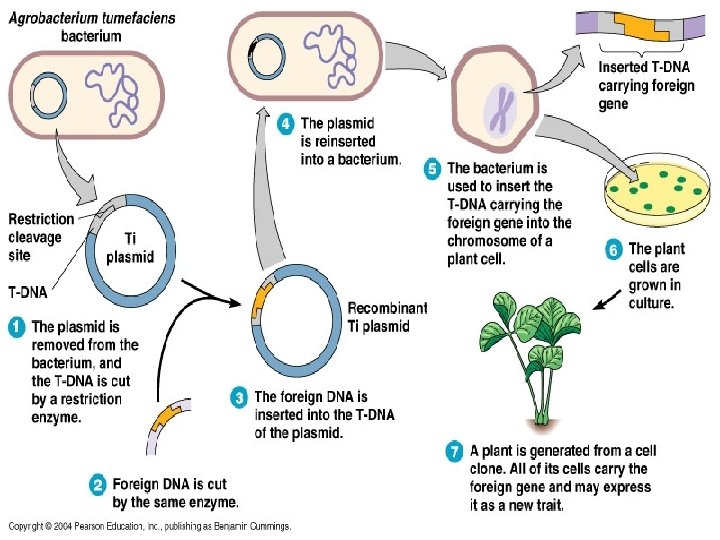
Plasmid Applications • The plasmids used in transformation typically have three important elements: • A cloning site (a place to insert foreign DNAs) • An origin of replication • A selectable marker gene (e. g. resistance to ampicillin)

Plasmid Applications


Plasmid DNA isolation • Isolation of plasmid DNA from bacterial cells is an essential step for many molecular biology procedures. • Many protocols for large- and small-scale isolation of plasmids have been published. • The plasmid purification procedures, unlike the procedures for purification of genomic DNA, should involve removal not only of protein, but also another major impurity: bacterial chromosomal DNA.

Overnight Culture Suspension • Pick a single colony and inoculate in 5 ml of LB (Luria-Bertani) containing 20 mg/l ampicilin • Incubate overnight at 37 o. C • Centrifuge 1. 5 ml of broth containing cells in a tube • Discard supernatant

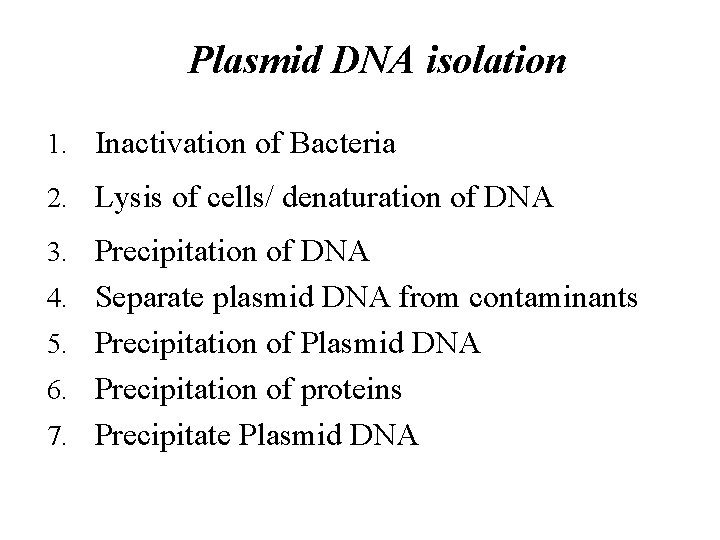
Plasmid DNA isolation 1. Inactivation of Bacteria 2. Lysis of cells/ denaturation of DNA 3. Precipitation of DNA 4. Separate plasmid DNA from contaminants 5. Precipitation of Plasmid DNA 6. Precipitation of proteins 7. Precipitate Plasmid DNA

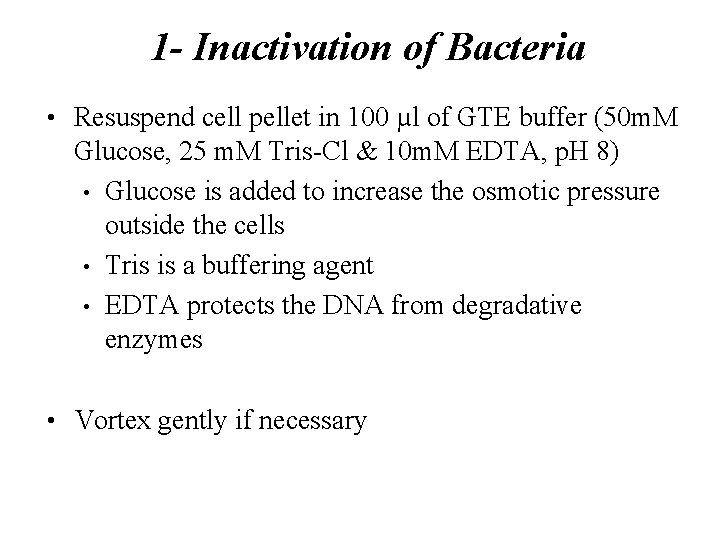
1 - Inactivation of Bacteria • Resuspend cell pellet in 100 µl of GTE buffer (50 m. M Glucose, 25 m. M Tris-Cl & 10 m. M EDTA, p. H 8) • Glucose is added to increase the osmotic pressure outside the cells • Tris is a buffering agent • EDTA protects the DNA from degradative enzymes • Vortex gently if necessary

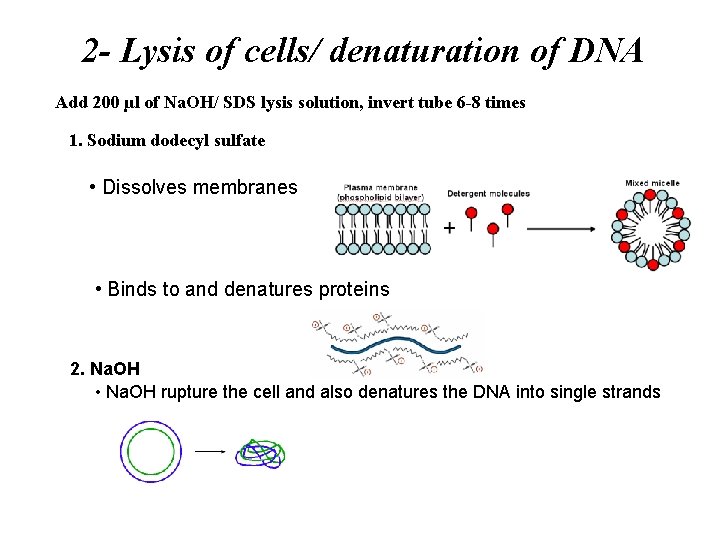
2 - Lysis of cells/ denaturation of DNA Add 200 µl of Na. OH/ SDS lysis solution, invert tube 6 -8 times 1. Sodium dodecyl sulfate • Dissolves membranes • Binds to and denatures proteins 2. Na. OH • Na. OH rupture the cell and also denatures the DNA into single strands

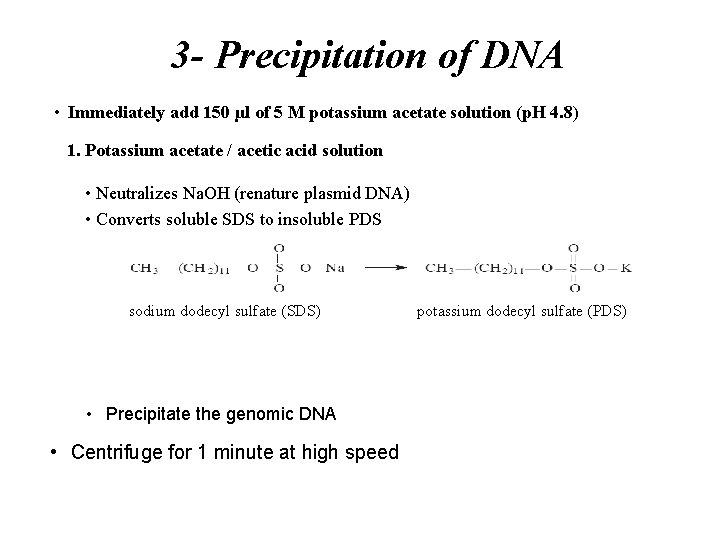
3 - Precipitation of DNA • Immediately add 150 µl of 5 M potassium acetate solution (p. H 4. 8) 1. Potassium acetate / acetic acid solution • Neutralizes Na. OH (renature plasmid DNA) • Converts soluble SDS to insoluble PDS sodium dodecyl sulfate (SDS) • Precipitate the genomic DNA • Centrifuge for 1 minute at high speed potassium dodecyl sulfate (PDS)

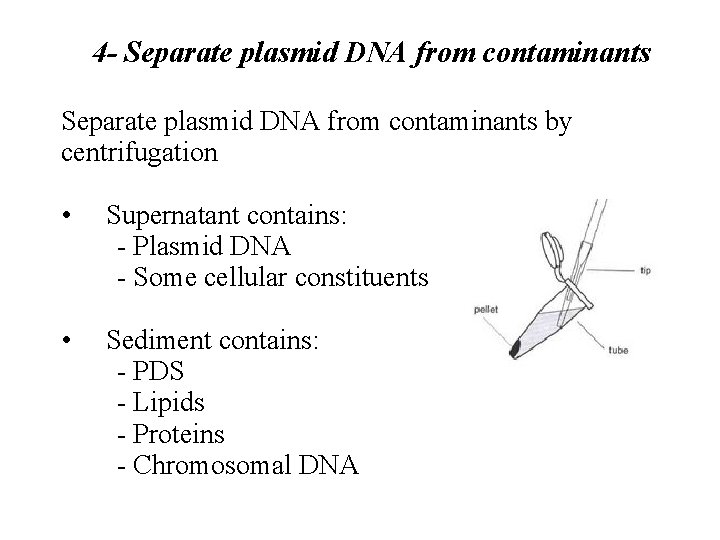
4 - Separate plasmid DNA from contaminants by centrifugation • Supernatant contains: - Plasmid DNA - Some cellular constituents • Sediment contains: - PDS - Lipids - Proteins - Chromosomal DNA

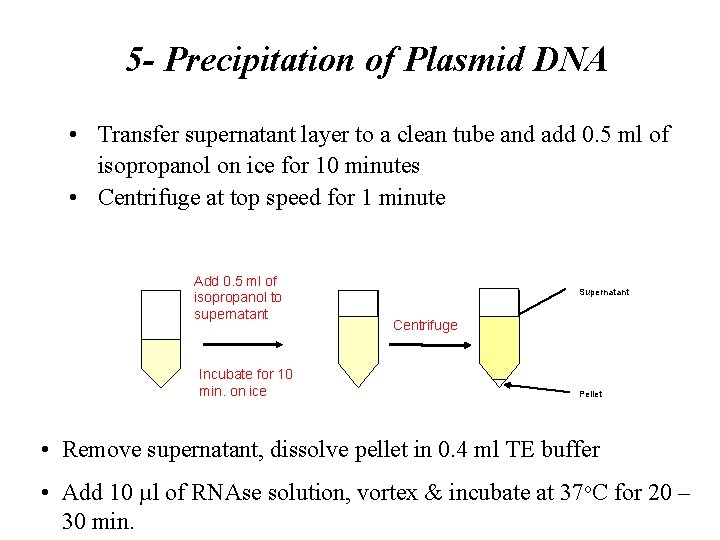
5 - Precipitation of Plasmid DNA • Transfer supernatant layer to a clean tube and add 0. 5 ml of isopropanol on ice for 10 minutes • Centrifuge at top speed for 1 minute Add 0. 5 ml of isopropanol to supernatant Incubate for 10 min. on ice Supernatant Centrifuge Pellet • Remove supernatant, dissolve pellet in 0. 4 ml TE buffer • Add 10 µl of RNAse solution, vortex & incubate at 37 o. C for 20 – 30 min.

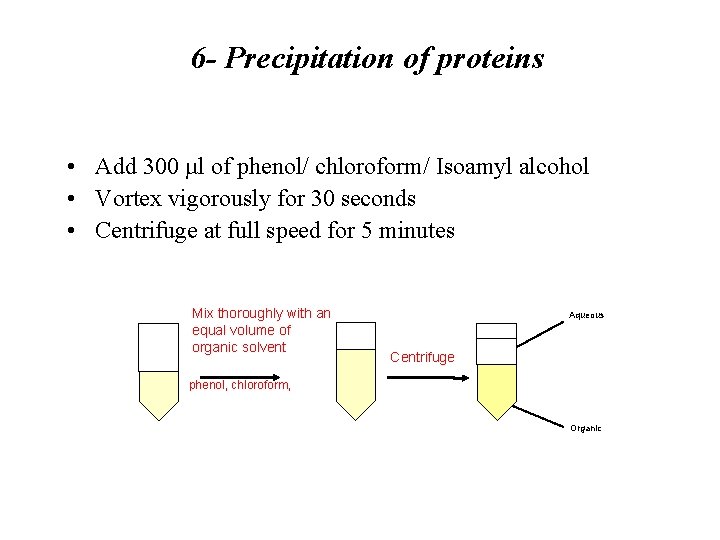
6 - Precipitation of proteins • Add 300 µl of phenol/ chloroform/ Isoamyl alcohol • Vortex vigorously for 30 seconds • Centrifuge at full speed for 5 minutes Mix thoroughly with an equal volume of organic solvent Aqueous Centrifuge phenol, chloroform, Organic

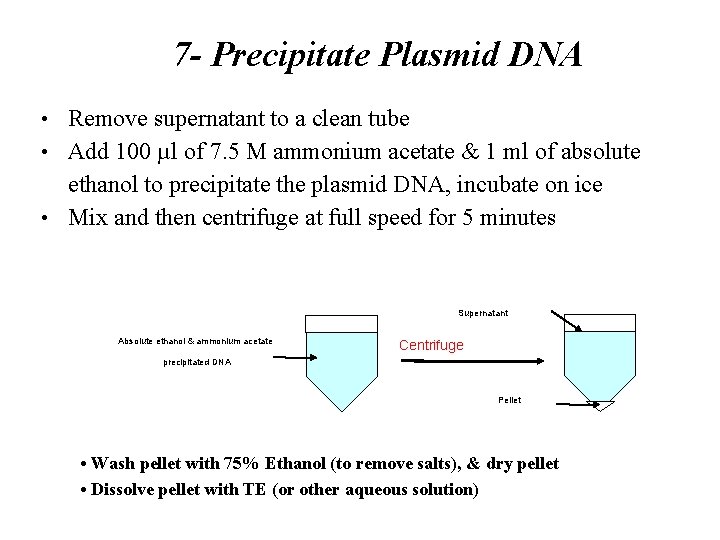
7 - Precipitate Plasmid DNA • Remove supernatant to a clean tube • Add 100 µl of 7. 5 M ammonium acetate & 1 ml of absolute ethanol to precipitate the plasmid DNA, incubate on ice • Mix and then centrifuge at full speed for 5 minutes Supernatant Absolute ethanol & ammonium acetate Centrifuge precipitated DNA Pellet • Wash pellet with 75% Ethanol (to remove salts), & dry pellet • Dissolve pellet with TE (or other aqueous solution)

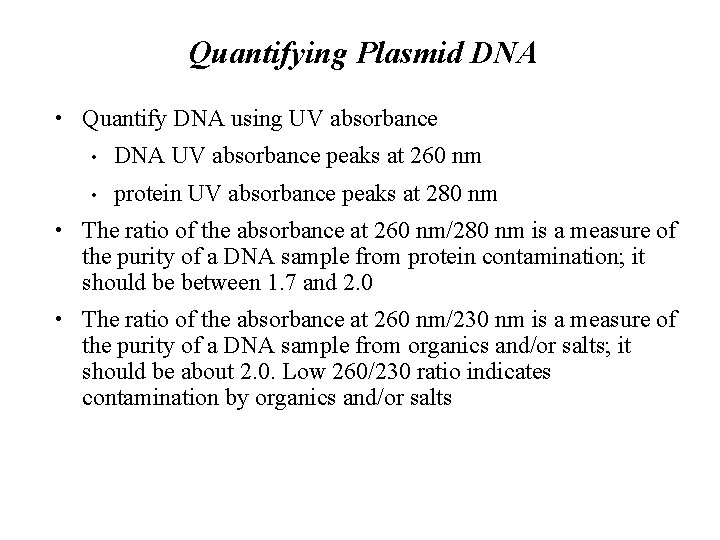
Quantifying Plasmid DNA • Quantify DNA using UV absorbance • DNA UV absorbance peaks at 260 nm • protein UV absorbance peaks at 280 nm • The ratio of the absorbance at 260 nm/280 nm is a measure of the purity of a DNA sample from protein contamination; it should be between 1. 7 and 2. 0 • The ratio of the absorbance at 260 nm/230 nm is a measure of the purity of a DNA sample from organics and/or salts; it should be about 2. 0. Low 260/230 ratio indicates contamination by organics and/or salts

• http: //www. dnatube. com/video/994/Plasmid • • • Isolation http: //www. dnalc. org/resources/animations/transf ormation 1. html http: //www. learnerstv. com/animation. p hp? ani=167&cat=biology http: //resources. jorum. ac. uk/xmlui/bitstream/hand le/123456789/13704/page 83. htm? sequence=86 http: //www. sumanasinc. com/webcontent/animatio ns/content/dna_library. swf http: //www. sumanasinc. com/webcontent/animatio ns/content/plasmidcloning. html
 Plasmid isolation principle
Plasmid isolation principle Application of plasmid
Application of plasmid Strategic goals tactical goals operational goals
Strategic goals tactical goals operational goals Strategic goals tactical goals operational goals
Strategic goals tactical goals operational goals Recombinant dna technology
Recombinant dna technology Fraction
Fraction Kiwi dna extraction lab
Kiwi dna extraction lab Dna extraction
Dna extraction Dna extraction
Dna extraction Dna extraction from kiwi fruit
Dna extraction from kiwi fruit Wheat germ dna factory
Wheat germ dna factory Wheat germ dna factory
Wheat germ dna factory Dna extraction
Dna extraction Chelex dna extraction advantages and disadvantages
Chelex dna extraction advantages and disadvantages Strawberry dna extraction materials
Strawberry dna extraction materials General goals and specific goals
General goals and specific goals Examples of generic goals and product-specific goals
Examples of generic goals and product-specific goals Dna polymerase function in dna replication
Dna polymerase function in dna replication Chapter 11 dna and genes
Chapter 11 dna and genes Bioflix activity dna replication lagging strand synthesis
Bioflix activity dna replication lagging strand synthesis