Extraction of Human DNA Experiment Goals Isolation of














- Slides: 25

Extraction of Human DNA

Experiment Goals • Isolation of genomic DNA from human blood • Analysis of isolated DNA using – Agarose gel electrophoresis – Spectrophotometry

What is a DNA? • DNA, also known as deoxyribonucleic acid, • A fundamental molecule found in all living things • Carries the genetic information in the cell • Contains instructions for our body cells to perform their specific functions • The sequence of nucleotides determines individual hereditary characteristics

What is a DNA? • Basic unit of information in DNA is the gene • Human beings have about 30, 000 gene • Size of organism’s genome is roughly a measure of its complexity • Viruses • E. coli • Human 5 -10 kb 4, 640 kb 2, 900, 000 kb

DNA Extraction • DNA extraction is a routine procedure to isolate & collect DNA. • DNA extraction is the first step for subsequent molecular or forensic analysis. • DNA can be extracted from almost any intact cellular tissue • • Skin, blood, saliva, semen, mucus, muscle tissue, bone marrow, etc.

Nucleic Acid Preparation Applications • Medical studies – Understanding genetic disorders at molecular level – Rapid detection of genetic disorders in a patient • Agricultural studies – Plant and animal breeding • Criminology/Paternity testing – DNA fingerprinting to identify individuals.

Basic steps in DNA extraction • There are three basic steps in a DNA extraction, the details of which may vary depending on the type of sample and any substances that may interfere with the extraction and subsequent analysis. – Break open cells and remove membrane lipids – Remove cellular and histone proteins bound to the DNA, by adding a protease, by precipitation with sodium or ammonium acetate, or by using a phenol/chloroform extraction step. – Precipitate DNA in cold ethanol or isopropanol, DNA is insoluble in alcohol and clings together, this step also removes salts.

Procedure

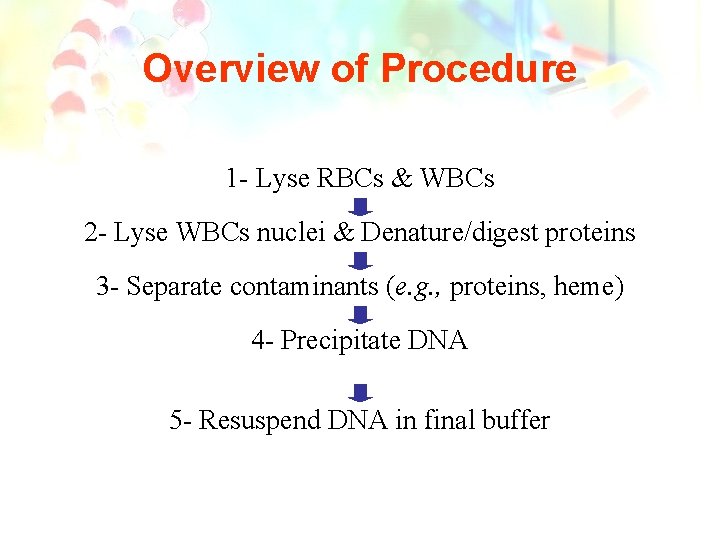
Overview of Procedure 1 - Lyse RBCs & WBCs 2 - Lyse WBCs nuclei & Denature/digest proteins 3 - Separate contaminants (e. g. , proteins, heme) 4 - Precipitate DNA 5 - Resuspend DNA in final buffer

Blood Collection • Blood collected in disodium EDTA tube • Samples can be stored at -20 o. C or -70 o. C • Fresh samples are kept in freezer for a few hours to facilitate RBCs hemolysis • Allow samples to thaw before starting the extraction

1 - RBCs Lysis • Pipette 3 mls of whole blood in a conical centrifuge tube • Add 9 mls of 1 X erythrocyte lysing buffer (0. 155 M NH 4 Cl, 10 m. M KHCO 3, 0. 1 m. M Na 2 EDTA, p. H 7. 4) • • • Leave 10 min. at RT, mix occasionally Centrifuge at 4000 rpm for 5 min Discard supernatent White pellet is observed at bottom of tube Wash pellet 3 times by adding 3 mls of buffer, incubate 10 min at RT, & centrifuge

2 - WBCs nuclei Lysis & proteins digestion • Add 1. 5 mls of SE buffer (75 m. M Na. Cl, 25 m. M Na 2 EDTA, p. H 8. 0) containing 100µg/ml of Proteinase K & 1% sodium dodecyl sulphate (SDS) to the pellet • Incubate at 37 -55 o. C overnight in a water bath or incubator • WBCs nuclei denatured & DNA goes out in solution

3 - Separate contaminants from DNA • After incubation add 1. 5 mls of SE buffer, 750 µl of 6 M Na. Cl & 3. 75 mls chloroform • Mix vigorously on vortex for 20 sec • Mix for 30 min (on rotator) • Centrifuge for 10 min at 2000 rpm • 2 phases are observed • DNA is extracted in supernatant & proteins in the lower phase • Transfer upper aqueous phase (containing DNA) to a clean tube

4 - Precipitate DNA • Add an equal volume of isopropanol • DNA will be precipitated by gentle swirling & observed as a white thread like strand • Using a sterile spatula or loop transfer the DNA strand into a sterile microcentrifuge tube containing 1 ml of 75% ethanol • Wash by inversion to remove any remaining salts • Centrifuge, discard supernatent • Repeat the washing step, then centrifuge • Remove supernatant, and dry the pellet

5 - Resuspend DNA in final buffer • Dried pellet is resuspended in TE buffer and left overnight on a rotator

DNA Analysis • Different methods for assessing quantity & quality of extracted DNA – Agarose gel electrophoresis – UV spectrophotometry

Checking the Quality of DNA • The product of DNA extracted will be used in subsequent experiments • Poor quality DNA will not perform well in PCR

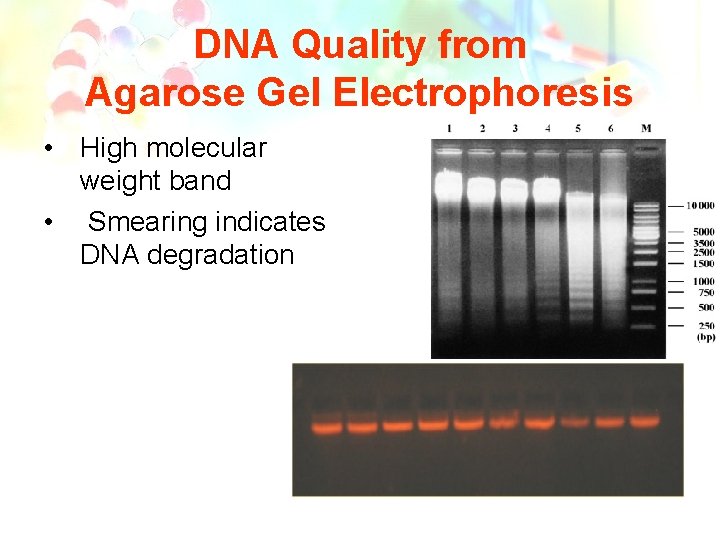
Quality from Agarose Gel Electrophoresis • Quality of DNA extracted is assessed using the following simple protocol: • • Mix 5 µL of DNA with 5 µL of loading Dye Load this mixture into a 1% agarose gel Stain with ethidium bromide Electrophorese at 70– 80 volts, 45– 90 minutes.

DNA Quality from Agarose Gel Electrophoresis • High molecular weight band • Smearing indicates DNA degradation

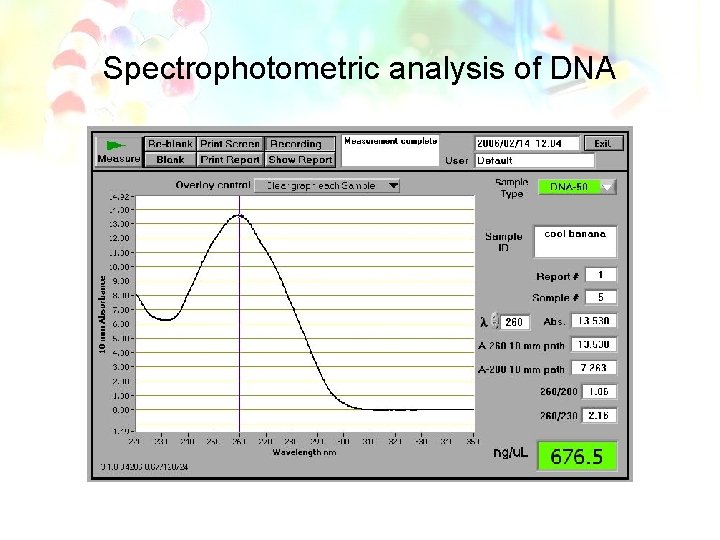
Nucleic Acid Characterization • Absorption Spectra – Absorb light in ultraviolet range, most strongly in the 254 -260 nm range • Useful for quantification of samples

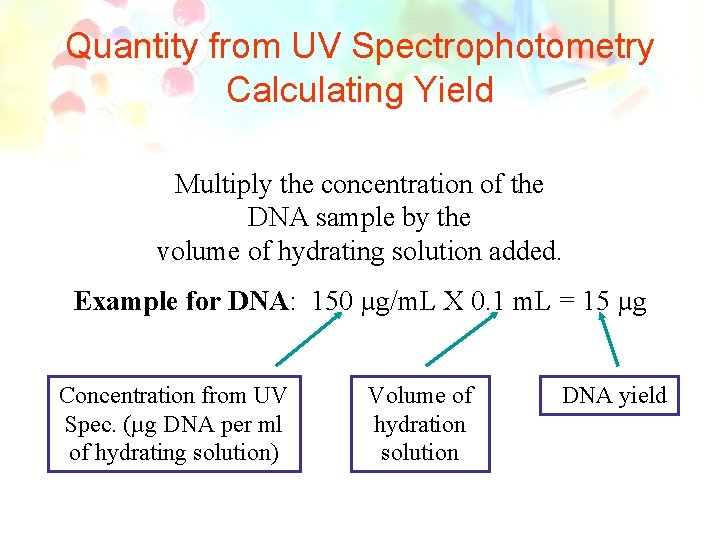
Quantity from UV Spectrophotometry Calculating Yield Multiply the concentration of the DNA sample by the volume of hydrating solution added. Example for DNA: 150 µg/m. L X 0. 1 m. L = 15 µg Concentration from UV Spec. (µg DNA per ml of hydrating solution) Volume of hydration solution DNA yield

Spectrophotometric analysis of DNA

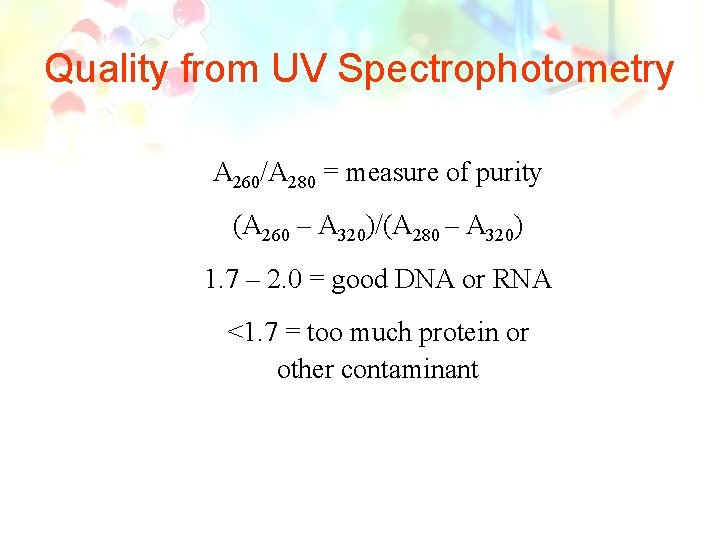
Quality from UV Spectrophotometry • DNA absorb maximally at 260 nm. • Proteins absorb at 280 nm. • Background scatter absorbs at 320 nm.

Quality from UV Spectrophotometry A 260/A 280 = measure of purity (A 260 – A 320)/(A 280 – A 320) 1. 7 – 2. 0 = good DNA or RNA <1. 7 = too much protein or other contaminant

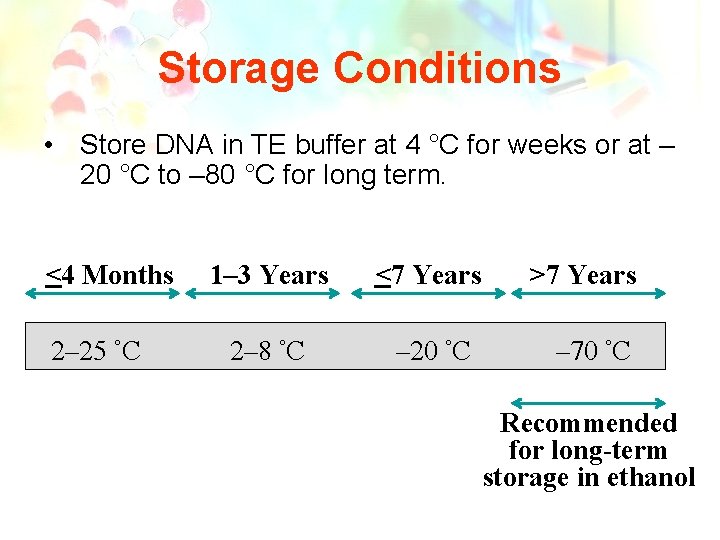
Storage Conditions • Store DNA in TE buffer at 4 °C for weeks or at – 20 °C to – 80 °C for long term. <4 Months 2– 25 °C 1– 3 Years <7 Years 2– 8 °C – 20 °C >7 Years – 70 °C Recommended for long-term storage in ethanol
 Strategic goals tactical goals operational goals
Strategic goals tactical goals operational goals Strategic goals tactical goals operational goals
Strategic goals tactical goals operational goals Strawberry dna extraction materials
Strawberry dna extraction materials Dna extraction
Dna extraction Wheat germ dna factory
Wheat germ dna factory Dna extraction
Dna extraction Difference between dna and rna extraction
Difference between dna and rna extraction Dna extraction
Dna extraction Dna extraction from wheat germ
Dna extraction from wheat germ Chelex dna extraction advantages and disadvantages
Chelex dna extraction advantages and disadvantages Kiwi dna extraction
Kiwi dna extraction Spooling dna definition
Spooling dna definition Principle of dna isolation
Principle of dna isolation General goals and specific goals
General goals and specific goals Examples of generic goals and product-specific goals
Examples of generic goals and product-specific goals Function of dna polymerase 3
Function of dna polymerase 3 Bioflix activity dna replication nucleotide pairing
Bioflix activity dna replication nucleotide pairing Coding dna and non coding dna
Coding dna and non coding dna Replication process
Replication process Chapter 11 dna and genes
Chapter 11 dna and genes Backwash effect ap human geography
Backwash effect ap human geography Cs 273
Cs 273 Replication of dna higher biology
Replication of dna higher biology Human vs non human bones
Human vs non human bones Chapter 8 human needs and human development
Chapter 8 human needs and human development Chapter 8 human needs and human development
Chapter 8 human needs and human development