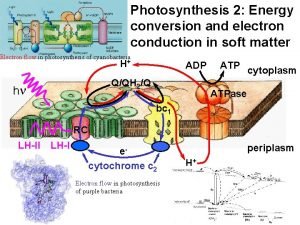
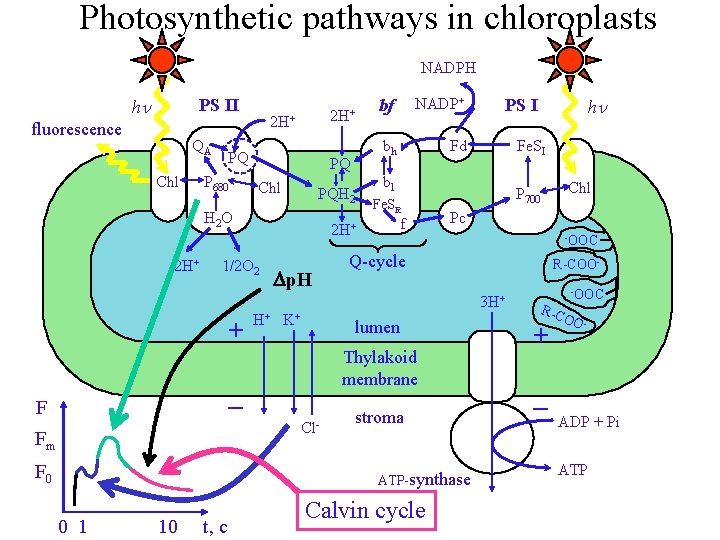
Photosynthetic pathways in chloroplasts NADPH hn PS II




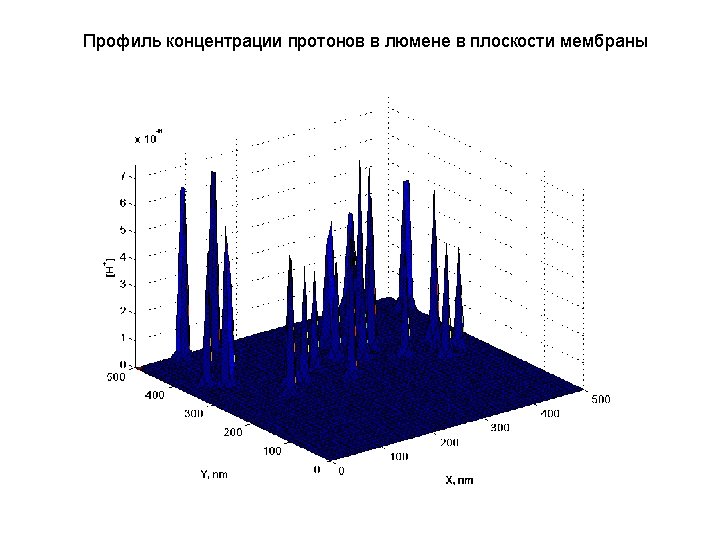
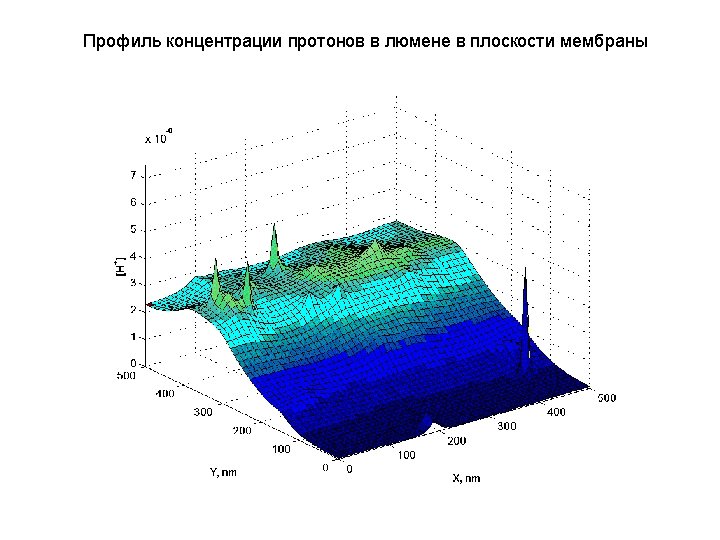
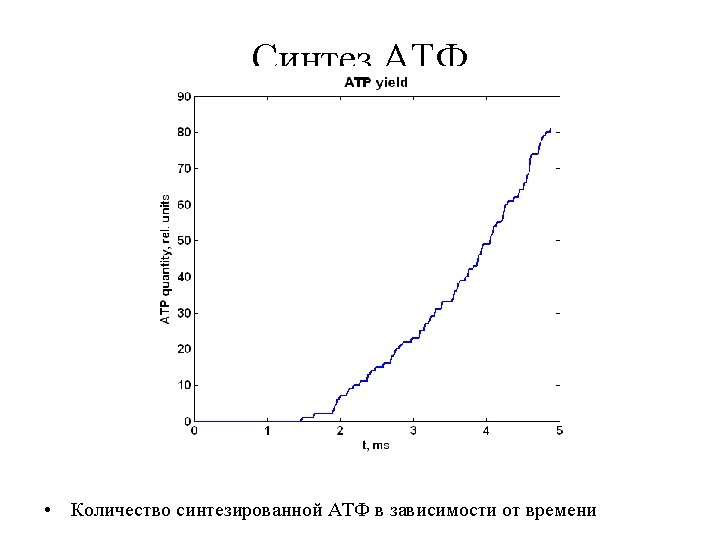
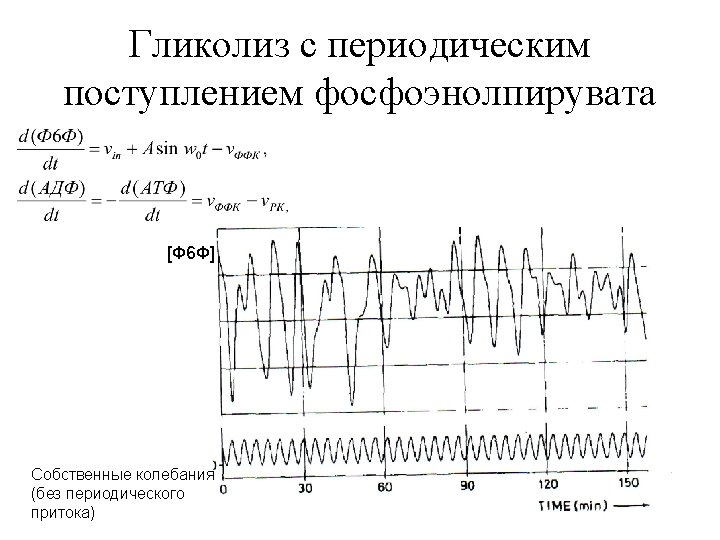
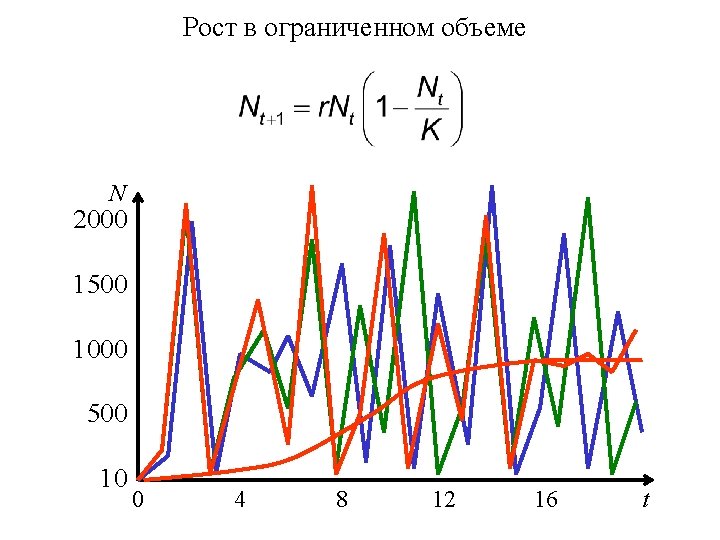
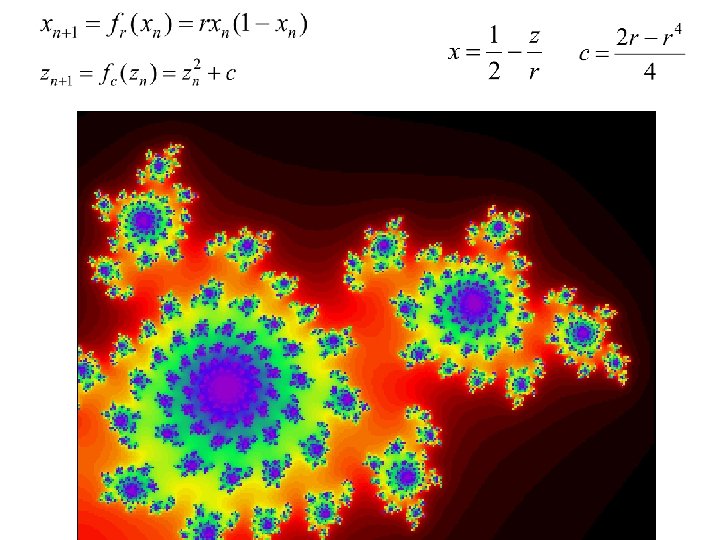
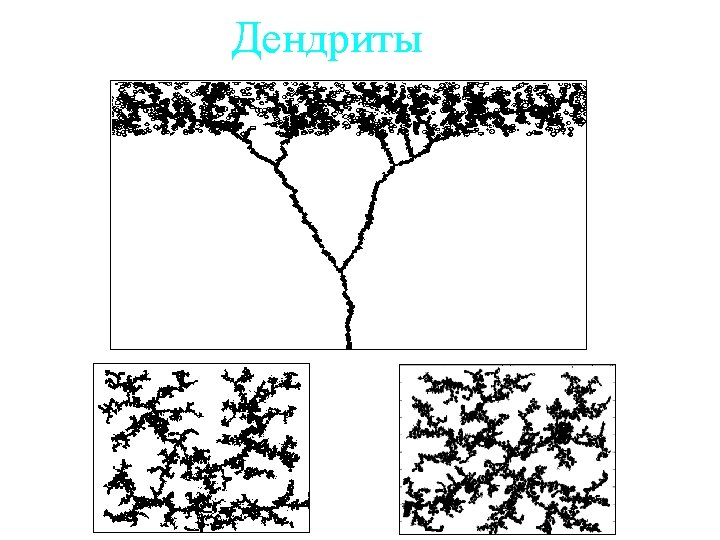


- Slides: 40




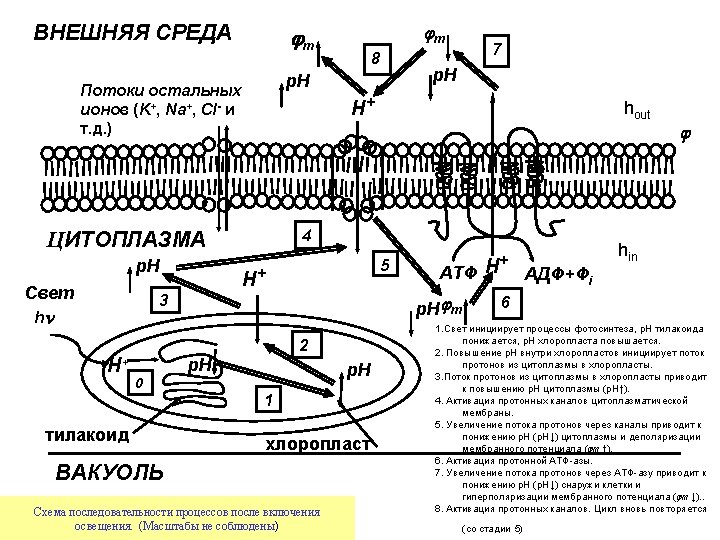
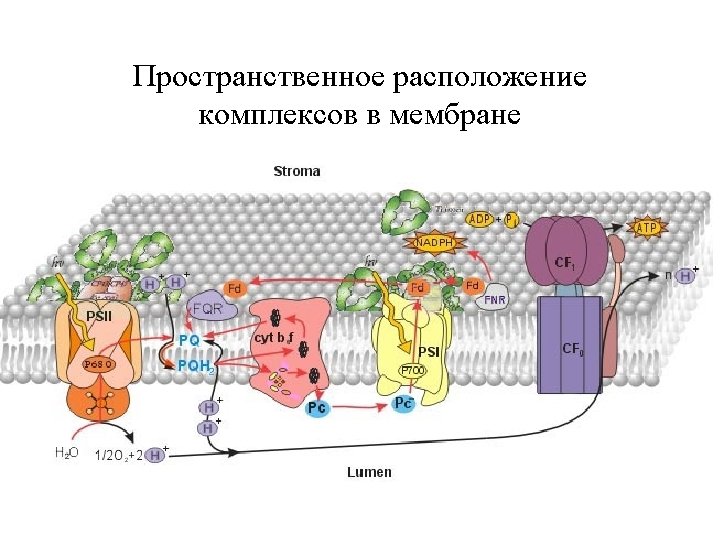
Photosynthetic pathways in chloroplasts NADPH hn PS II fluorescence QA Chl 2 H+ PQ P 680 PQ Chl PQH 2 H 2 O 2 H+ 1/2 O 2 Dp. H bf NADP+ bh Fd Fe. SI bl Fe. SR f + Pc -OOC Q-cycle lumen Thylakoid membrane _ F Cl- Fm F 0 stroma ATP-synthase 0 1 10 t, c Chl P 700 R-COO- 3 H+ H+ K+ hn PS I Calvin cycle R-C -OOC + OO - _ ADP + Pi ATP

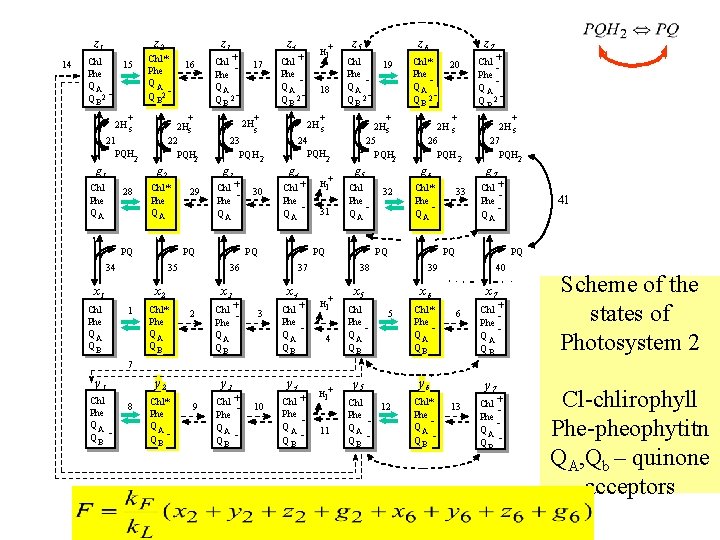
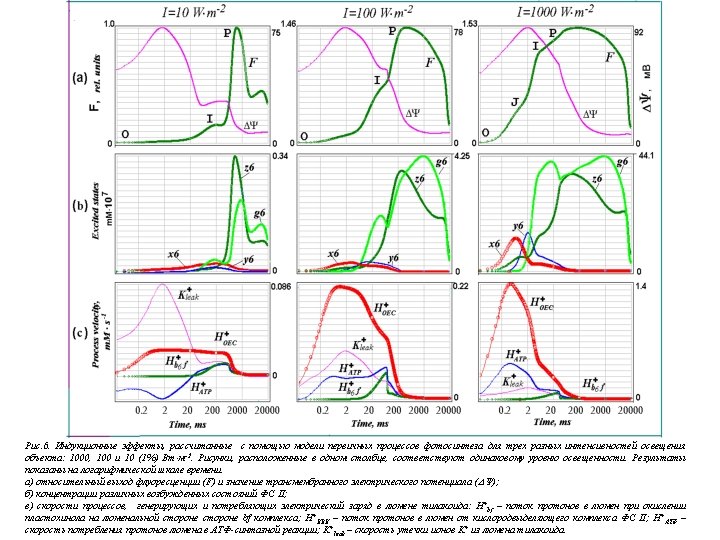
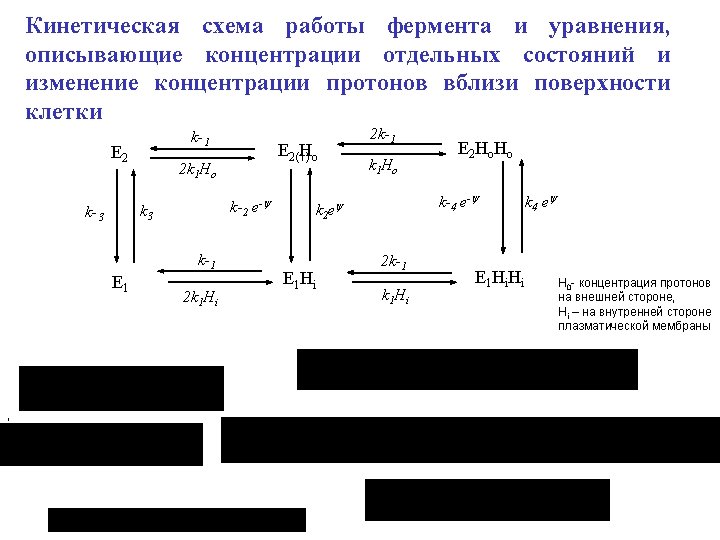
z 1 14 z 2 Chl* Phe QA Q B 2 - Chl 15 Phe QA Q B 2 + 2 H s 2 g 1 g 2 28 Chl* Phe QA 29 35 z 4 17 Chl* Phe QA QB Chl Phe Q A Q B 2 - + 2 H s Chl Phe QA 30 Chl Phe Q A PQ Chl + Phe QA QB Hl + 31 g 5 Chl Phe Q A PQ 3 Chl + Phe Q A QB z 7 Chl* Phe Q A Q B 2 - g 6 32 l 4 + 2 H s 27 PQH 2 g 7 Chl* Phe Q A 33 Chl + Phe Q A 40 x 6 5 Chl* Phe Q A QB 41 PQ 39 x 5 Chl Phe Q A QB Chl + Phe Q A Q B 2 - PQ 38 H+ 20 + 2 H s 26 PQH 2 PQ 37 x 4 19 + 2 Hs 25 PQH 2 2 + z 6 Chl Phe Q A Q B 2 - 24 PQH g 4 x 3 2 18 z 5 + 2 H s 23 PQH 2 + Hl + + 36 x 2 1 Chl Phe QA Q B 2 - PQ 34 x 1 + g 3 PQ Chl Phe QA QB 16 + 2 Hs 22 PQH 2 21 PQH Chl Phe QA z 3 x 7 6 Chl + Phe Q A QB Scheme of the states of Photosystem 2 7 y 1 Chl Phe QA Q B - y 2 8 Chl* Phe QA Q B - y 3 9 + Chl Phe QA Q B - y 4 10 + Chl Phe Q A Q B Hl + 11 y 5 Chl 12 Phe Q A Q B y 6 Chl* Phe Q A Q B - y 7 13 Chl + Phe Q A Q B - Cl-chlirophyll Phe-pheophytitn QA, Qb – quinone acceptors

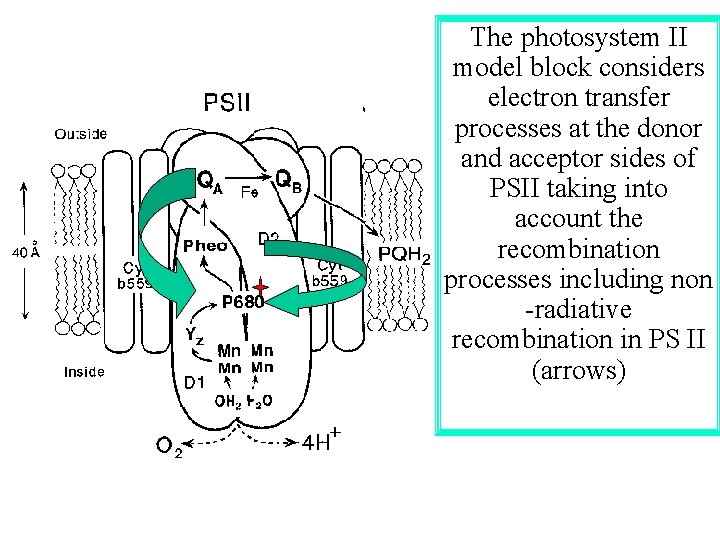
The photosystem II model block considers electron transfer processes at the donor and acceptor sides of PSII taking into account the recombination processes including non -radiative recombination in PS II (arrows)

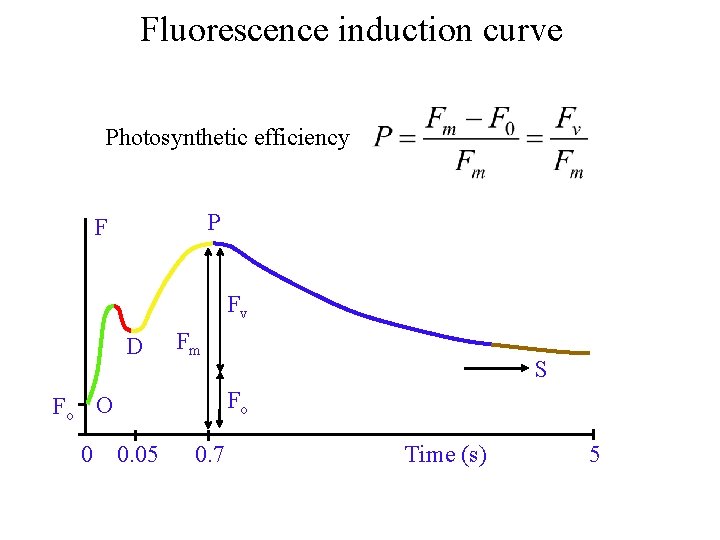
Fluorescence induction curve Photosynthetic efficiency P F Fv D Fo O Fm S Fo 0 0. 05 0. 7 Time (s) 5











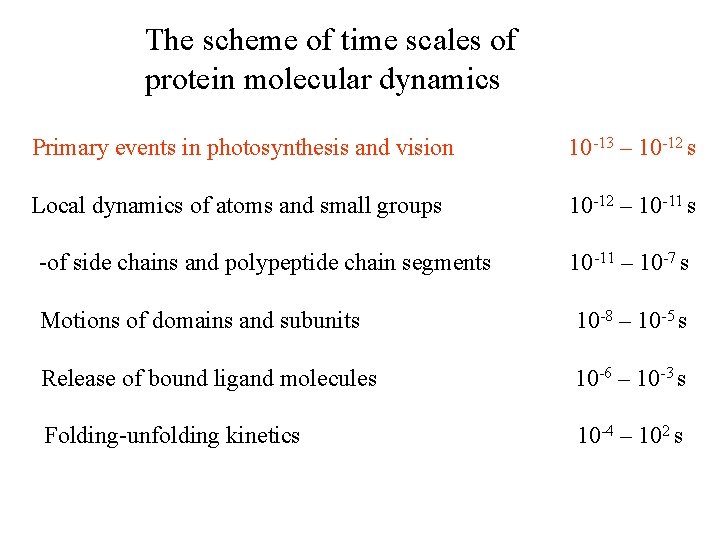
The scheme of time scales of protein molecular dynamics Primary events in photosynthesis and vision 10 -13 – 10 -12 s Local dynamics of atoms and small groups 10 -12 – 10 -11 s -of side chains and polypeptide chain segments 10 -11 – 10 -7 s Motions of domains and subunits 10 -8 – 10 -5 s Release of bound ligand molecules 10 -6 – 10 -3 s Folding-unfolding kinetics 10 -4 – 102 s

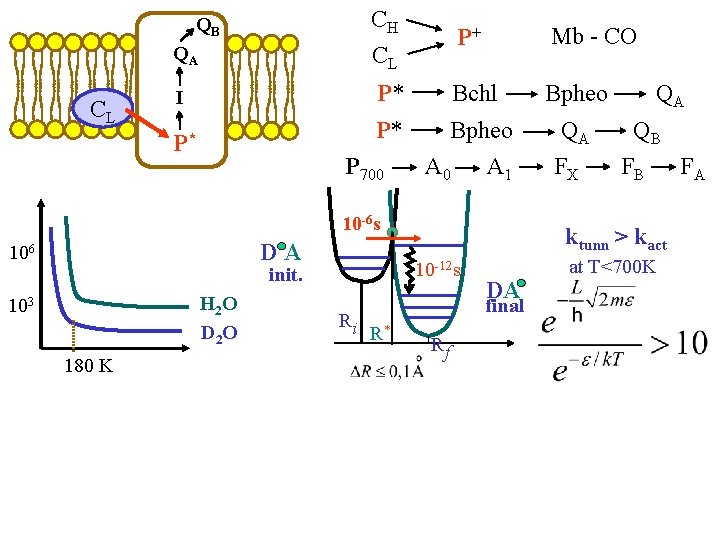
CH CL QB QA CL Mb - CO P+ I P* Bchl Bpheo QA P* P* Bpheo QA QB P 700 A 0 A 1 FX FB FA 10 -6 s DA 106 10 -12 s init. H 2 O D 2 O 103 180 K ktunn > kact Ri DA final R* Rf at T<700 K

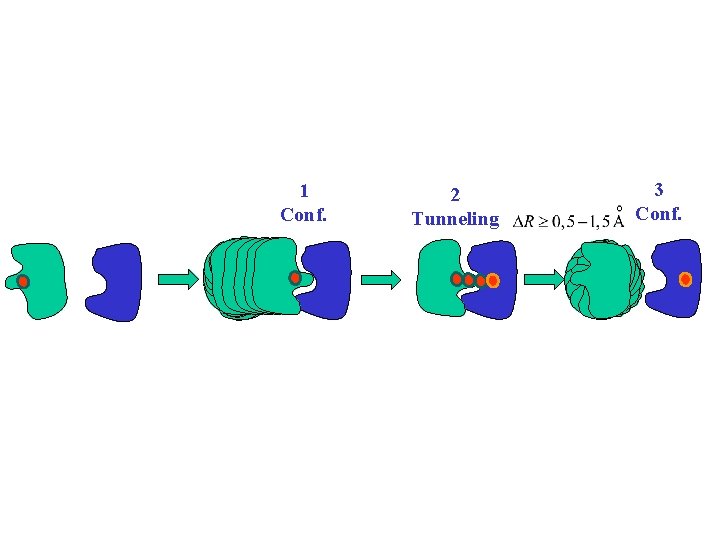
1 Conf. 2 Tunneling 3 Conf.

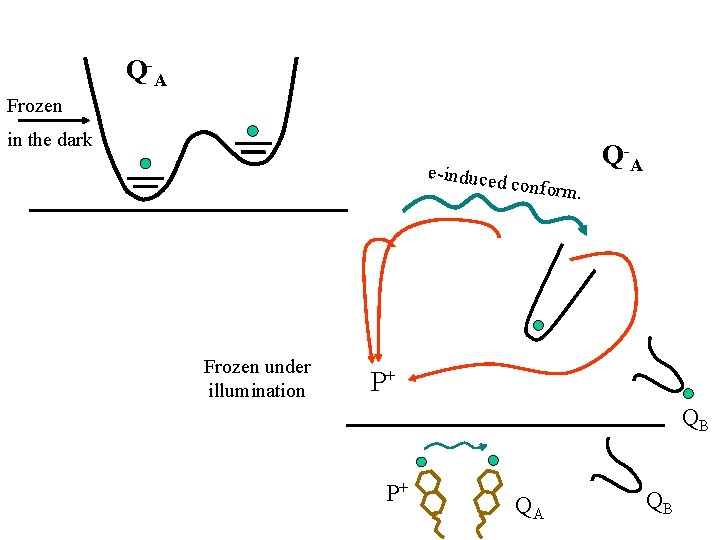
Q -A Frozen in the dark e-induced Frozen under illumination conform Q -A. P+ QB P+ QA QB


Scene of the direct model

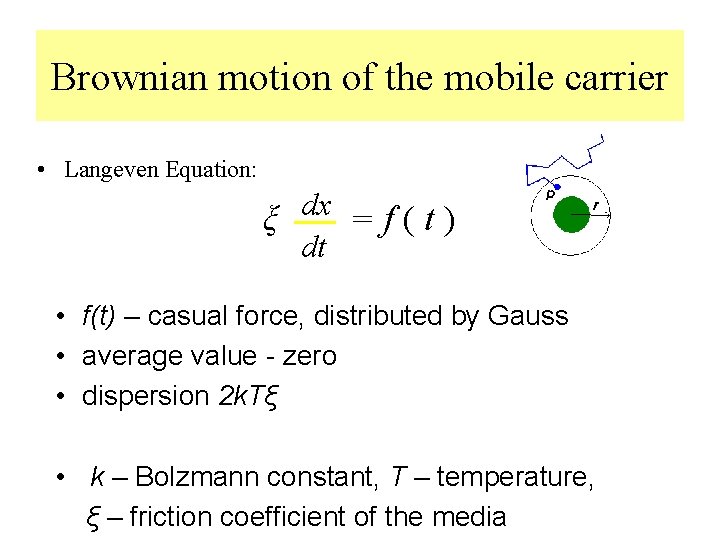
Brownian motion of the mobile carrier • Langeven Equation: dx ξ = f ( t ) dt • f(t) – casual force, distributed by Gauss • average value - zero • dispersion 2 k. Tξ • k – Bolzmann constant, T – temperature, ξ – friction coefficient of the media

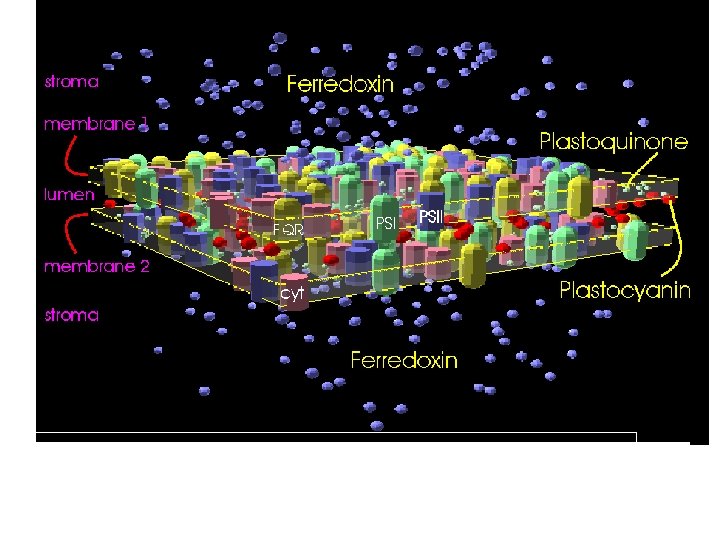
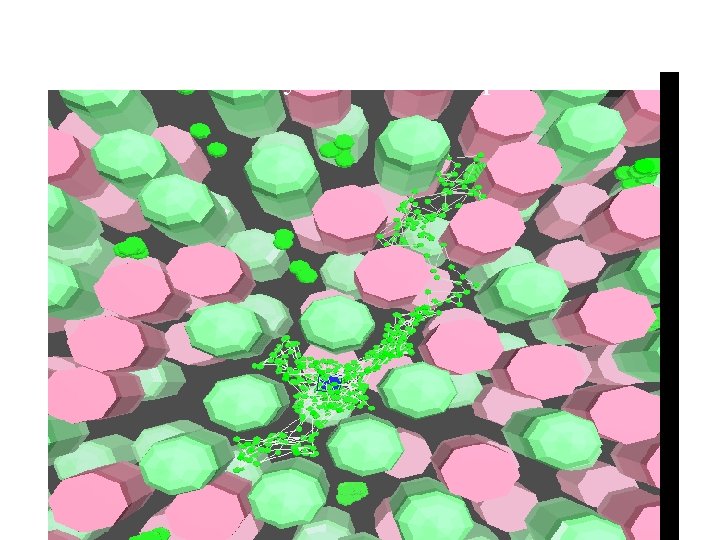
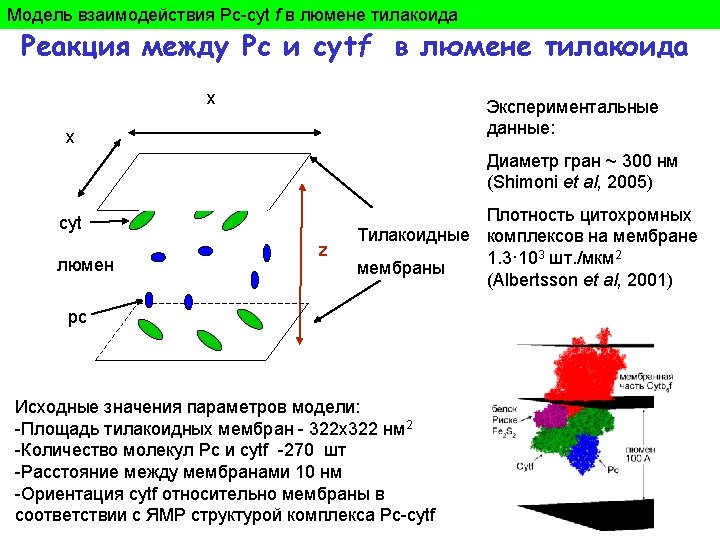
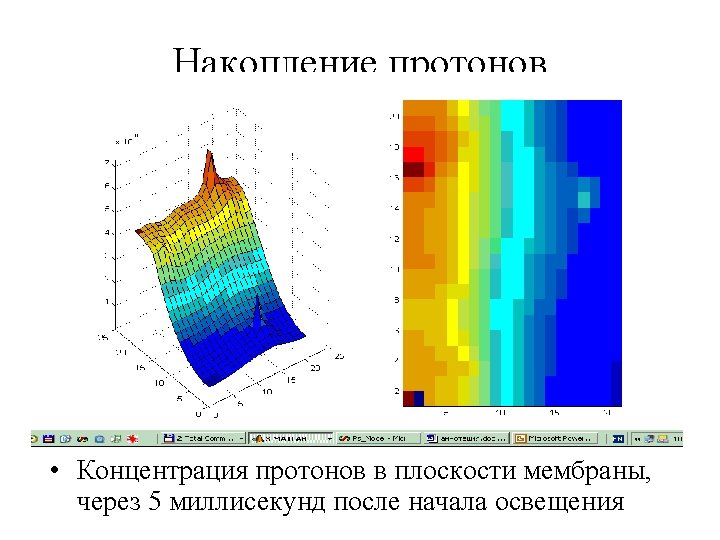
Model trajectory of PQ in membrane filled by PS 1 and cytochrome complexes

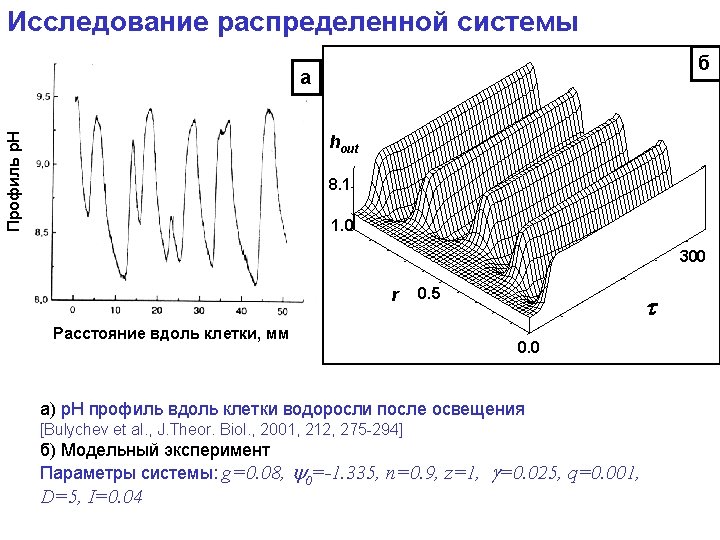
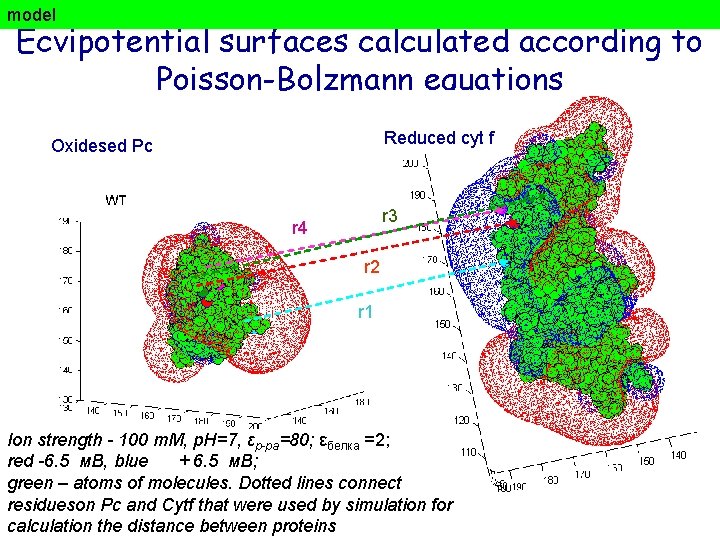
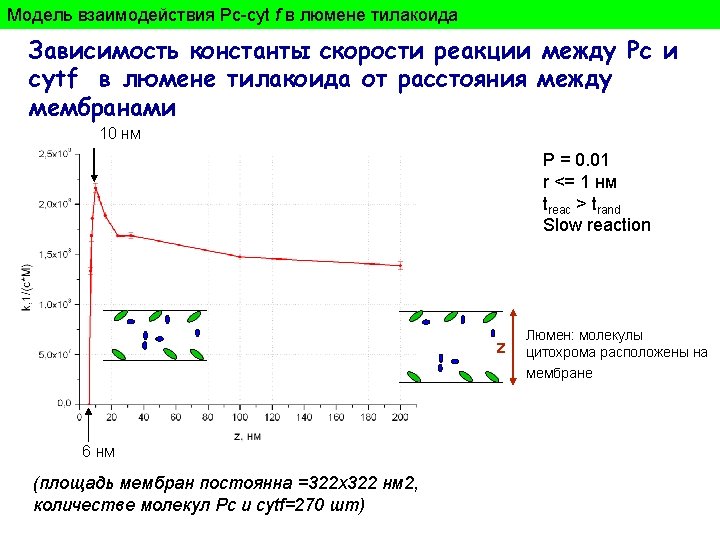
model Ecvipotential surfaces calculated according to Poisson-Bolzmann equations Reduced cyt f Oxidesed Рс r 3 r 4 r 2 r 1 Ion strength - 100 m. M, p. H=7, εр-ра=80; εбелка =2; red -6. 5 м. В, blue + 6. 5 м. В; green – atoms of molecules. Dotted lines connect residueson Pc and Cytf that were used by simulation for calculation the distance between proteins


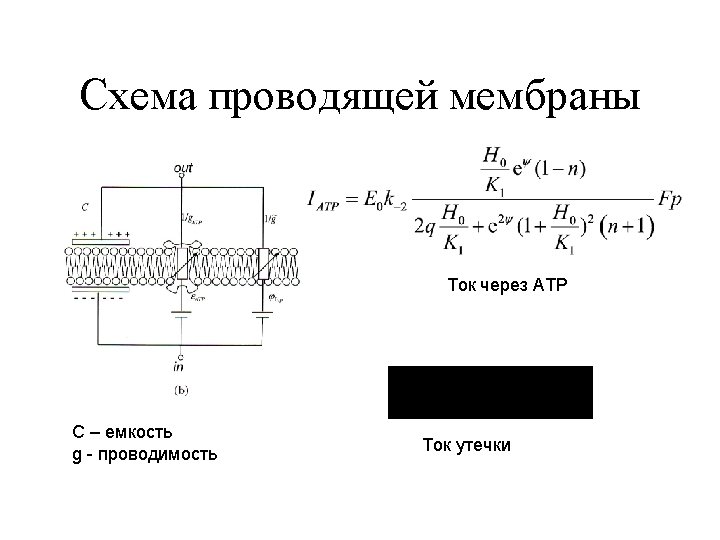
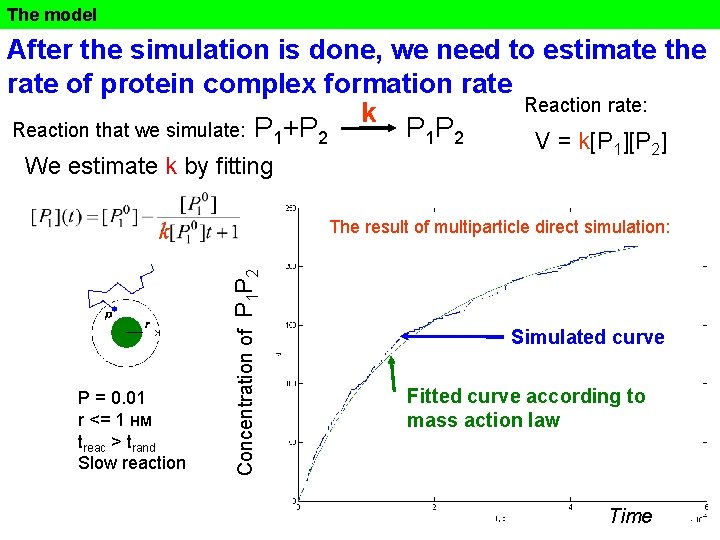
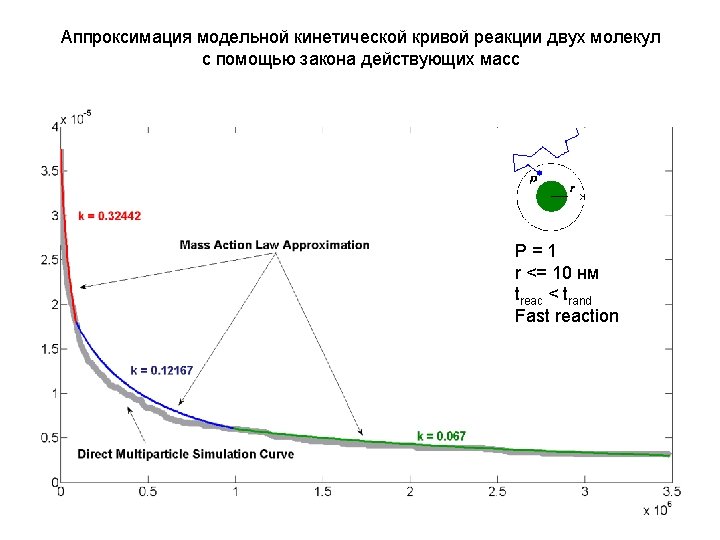
The model After the simulation is done, we need to estimate the rate of protein complex formation rate Reaction rate: k Reaction that we simulate: P 1+P 2 P 1 P 2 V = k[P ] We estimate k by fitting 1 2 The result of multiparticle direct simulation: P = 0. 01 r <= 1 нм treac > trand Slow reaction Concentration of P 1 P 2 k Simulated curve Fitted curve according to mass action law Time












• • • Galina Riznichenko Evgeny Grachev Natalia Beljaeva Pavel Gromov Ilia Kovalenko Dmitry Ustinin Anna Abaturova Tatjana Plusnina Nastja Lavrova Vladimir Paschenko Petr Noks
 Anabolismp
Anabolismp 6 fosfoglukonolakton
6 fosfoglukonolakton Pgal sentezi
Pgal sentezi Nadh vs nadph
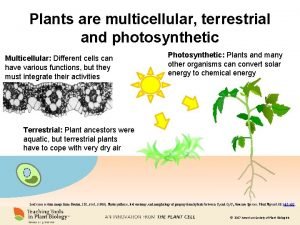
Nadh vs nadph Photosynthetic multicellular and terrestrial kingdom
Photosynthetic multicellular and terrestrial kingdom Photosynthetic phytoplankton
Photosynthetic phytoplankton Chloroplast contain saclike photosynthetic membranes called
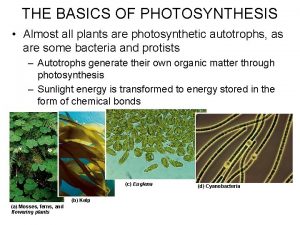
Chloroplast contain saclike photosynthetic membranes called Photosynthetic autotroph
Photosynthetic autotroph What is a photosynthetic organism
What is a photosynthetic organism Tiny, threadlike photosynthetic organisms
Tiny, threadlike photosynthetic organisms Cell
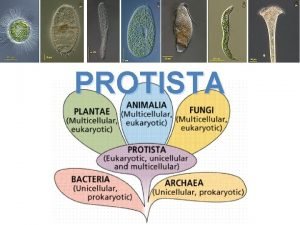
Cell Which kingdoms have photosynthetic organisms? *
Which kingdoms have photosynthetic organisms? * Volvox and paramecium
Volvox and paramecium Batang dikotil
Batang dikotil What kingdom is photosynthetic aquatic and unicellular
What kingdom is photosynthetic aquatic and unicellular Photosynthetic apparatus
Photosynthetic apparatus Photosynthetic cells
Photosynthetic cells Which two protists contain chloroplasts
Which two protists contain chloroplasts Mitochondria ppt slides
Mitochondria ppt slides A cell with numerous ribosomes is probably specialized for
A cell with numerous ribosomes is probably specialized for Mitochondria function
Mitochondria function Killing chloroplasts case study answers 5
Killing chloroplasts case study answers 5 Rough endoplasmic reticulum function
Rough endoplasmic reticulum function Vascular tissue
Vascular tissue Hybrid pathways
Hybrid pathways Pathways madison wi
Pathways madison wi Higher biology unit 2
Higher biology unit 2 Lambert high school pathways
Lambert high school pathways Ruth binks inverclyde council
Ruth binks inverclyde council Pathways to graduation
Pathways to graduation Metabolic pathways higher human biology
Metabolic pathways higher human biology Mt sac guided pathways
Mt sac guided pathways Exploitation plan example
Exploitation plan example Arithmetic reasoning
Arithmetic reasoning Sas curriculum pathways
Sas curriculum pathways Referral pathways
Referral pathways Thank you diversity
Thank you diversity Label the components of the conduction system.
Label the components of the conduction system. What are the 3 pathways to paternity?
What are the 3 pathways to paternity? Kde cte pathways
Kde cte pathways The pathways to new ventures for entrepreneurs
The pathways to new ventures for entrepreneurs