Napovedovanje imunskega odziva iz peptidnih mikromre Mitja Lutrek










- Slides: 75

Napovedovanje imunskega odziva iz peptidnih mikromrež Mitja Luštrek 1 (2), Peter Lorenz 2, Felix Steinbeck 2, Georg Füllen 2, Hans-Jürgen Thiesen 2 1 Odsek za inteligentne sisteme, Institut Jožef Stefan 2 Univerza v Rostocku

1. Introduction 2. Immune response prediction 3. Interpretation

1. Introduction 2. Immune response prediction 3. Interpretation

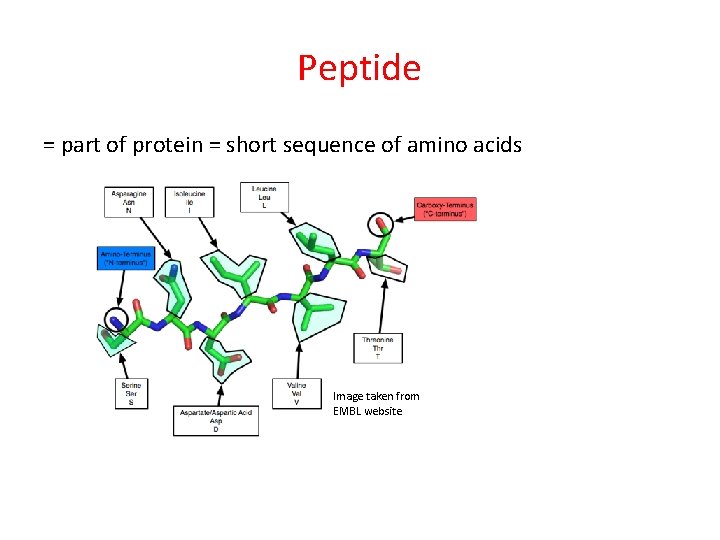
Peptide = part of protein = short sequence of amino acids Image taken from EMBL website

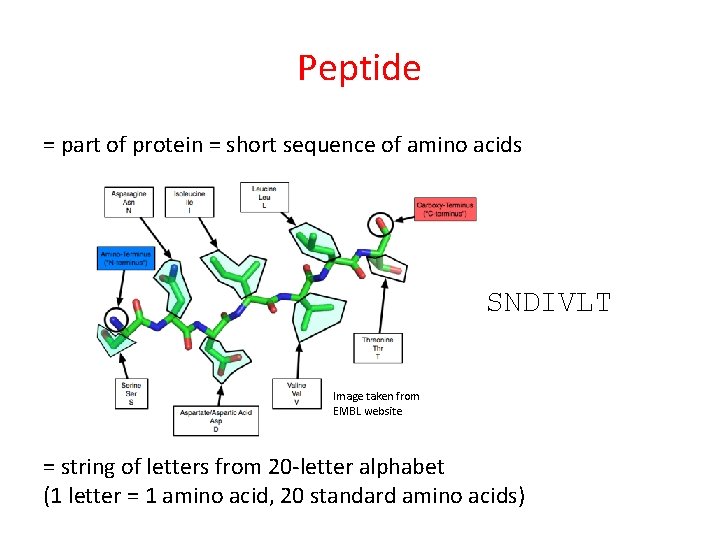
Peptide = part of protein = short sequence of amino acids SNDIVLT Image taken from EMBL website = string of letters from 20 -letter alphabet (1 letter = 1 amino acid, 20 standard amino acids)

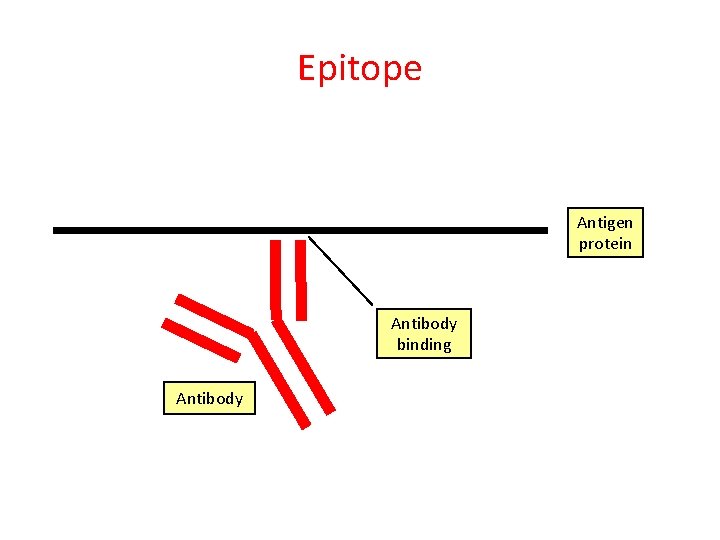
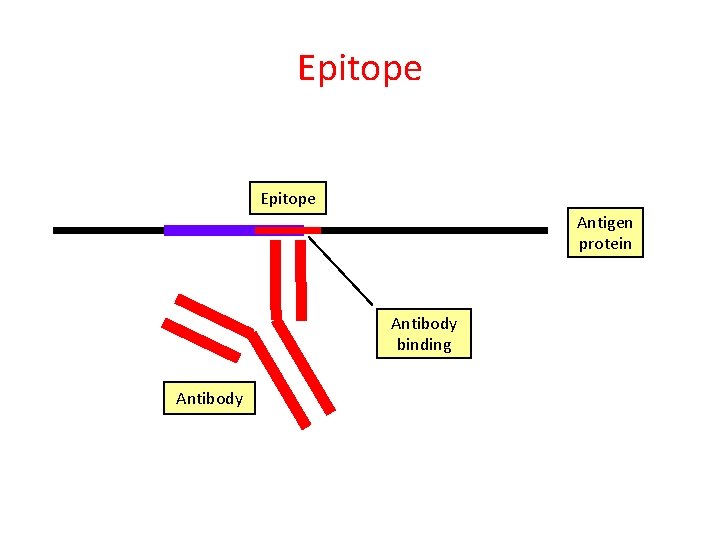
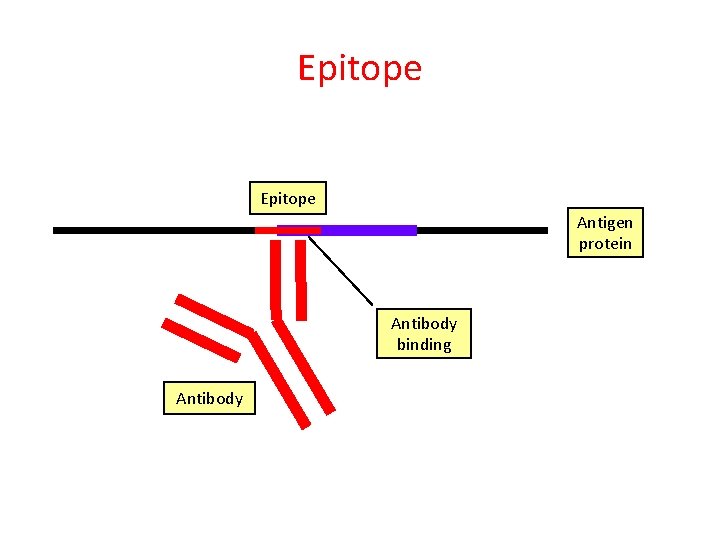
Epitope Antigen protein Antibody binding Antibody

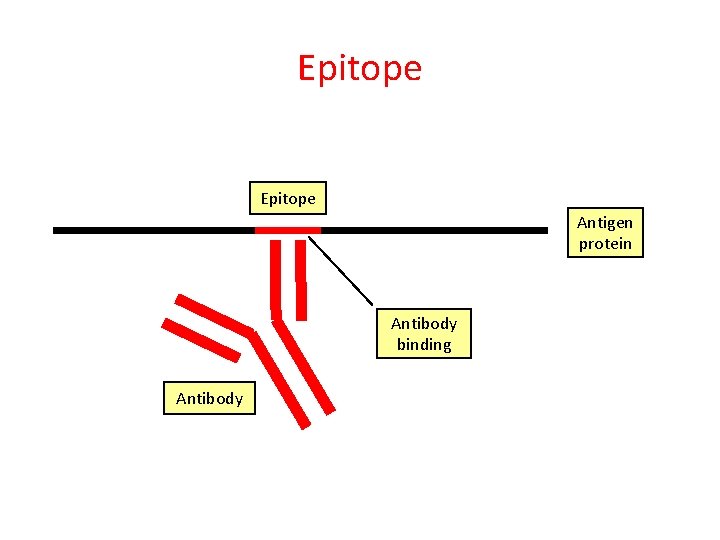
Epitope Antigen protein Antibody binding Antibody

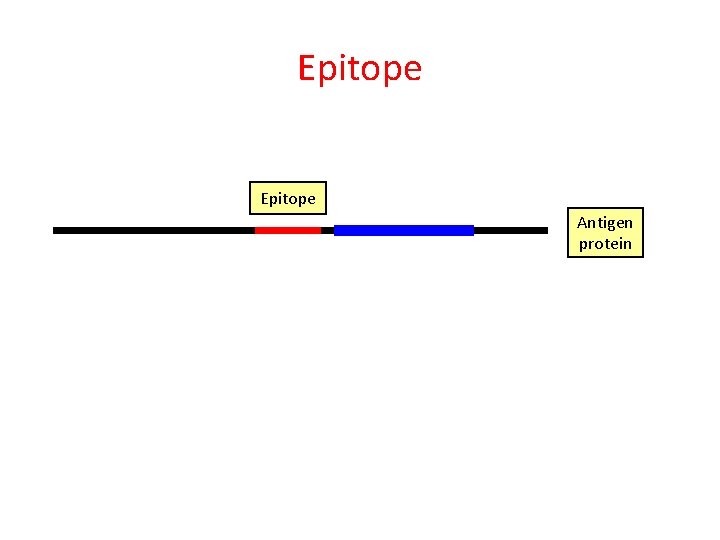
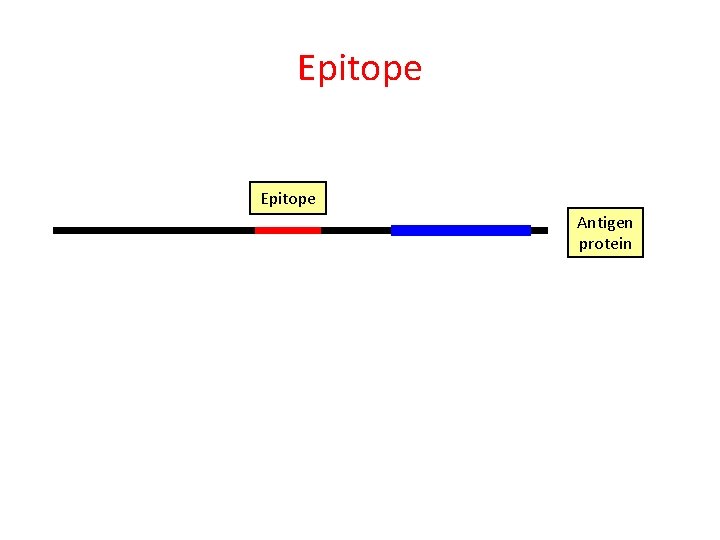
Epitope Peptide Antigen protein

Epitope Antigen protein

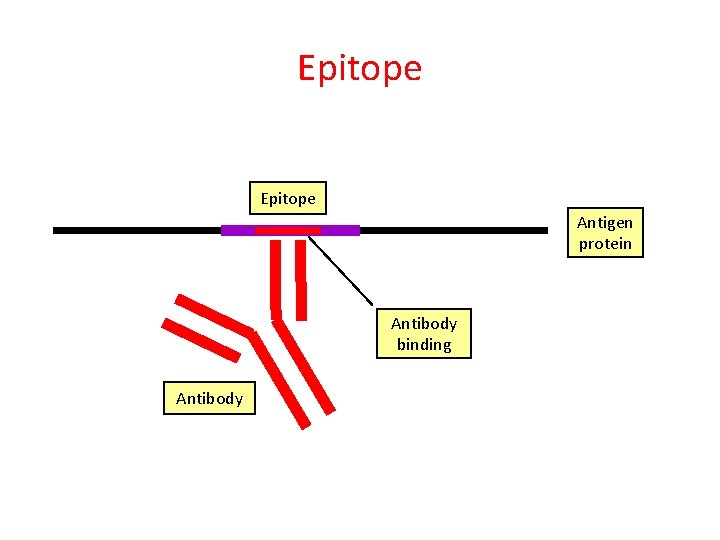
Epitope Antigen protein Antibody binding Antibody

Epitope Antigen protein Antibody binding Antibody

Epitope Antigen protein Antibody binding Antibody

Epitope Antigen protein

Epitope Antigen protein

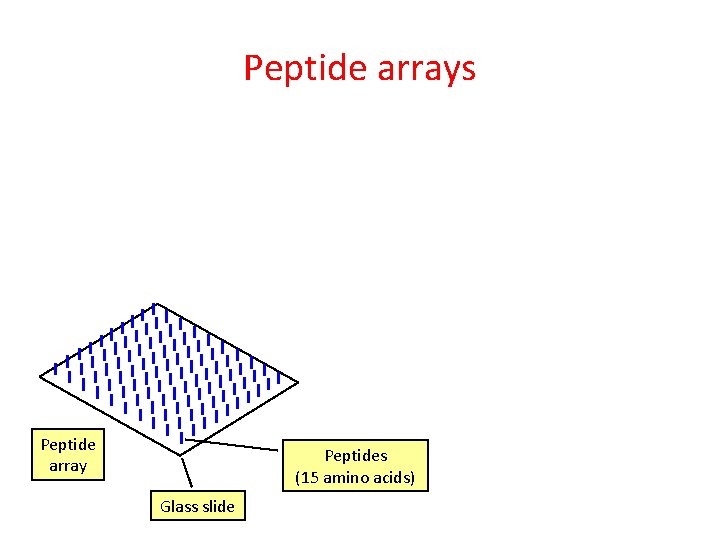
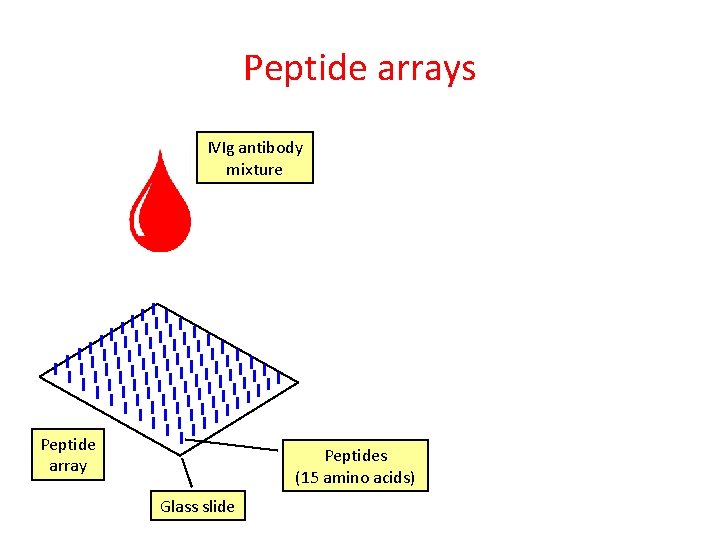
Peptide arrays Peptide array Peptides (15 amino acids) Glass slide

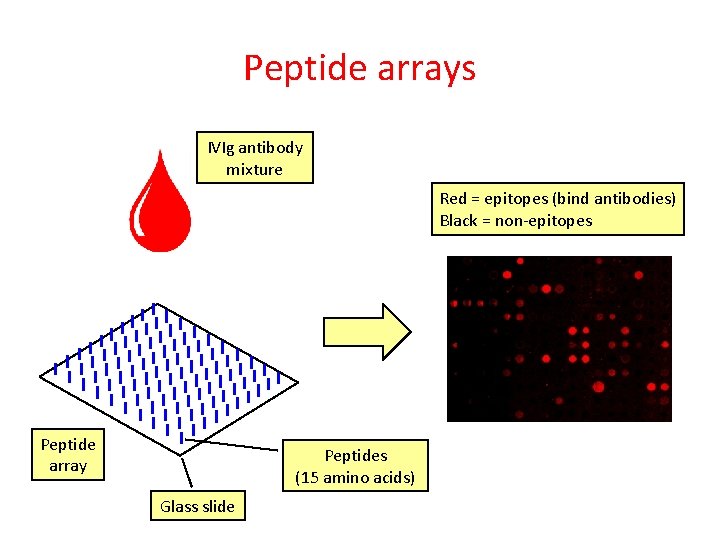
Peptide arrays IVIg antibody mixture Peptide array Peptides (15 amino acids) Glass slide

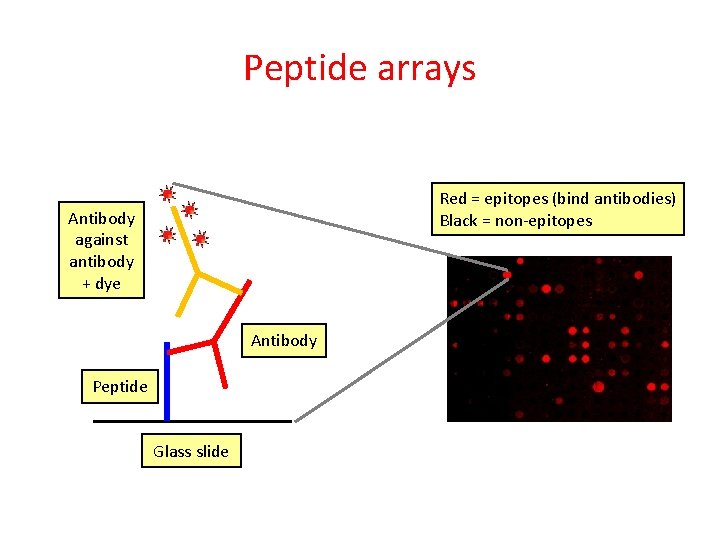
Peptide arrays IVIg antibody mixture Red = epitopes (bind antibodies) Black = non-epitopes Peptide array Peptides (15 amino acids) Glass slide

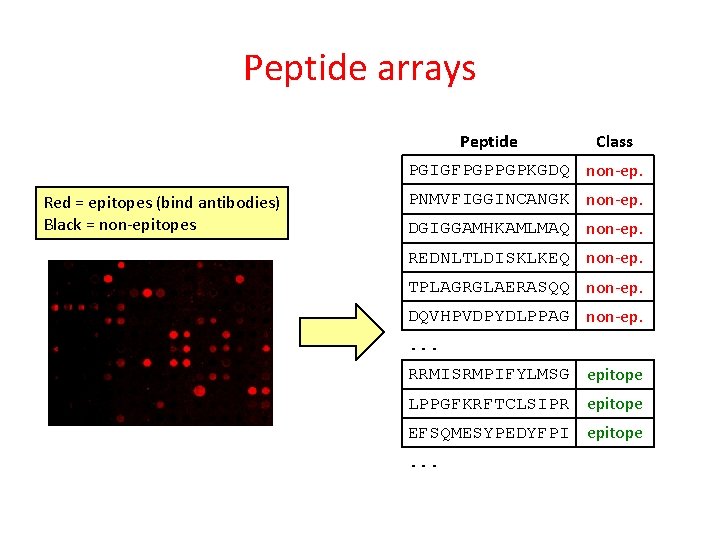
Peptide arrays Red = epitopes (bind antibodies) Black = non-epitopes Antibody against antibody + dye Antibody Peptide Glass slide

Peptide arrays Peptide Class PGIGFPGPPGPKGDQ non-ep. Red = epitopes (bind antibodies) Black = non-epitopes PNMVFIGGINCANGK non-ep. DGIGGAMHKAMLMAQ non-ep. REDNLTLDISKLKEQ non-ep. TPLAGRGLAERASQQ non-ep. DQVHPVDPYDLPPAG non-ep. . RRMISRMPIFYLMSG epitope LPPGFKRFTCLSIPR epitope EFSQMESYPEDYFPI epitope. . .

1. Introduction 2. Immune response prediction 3. Interpretation

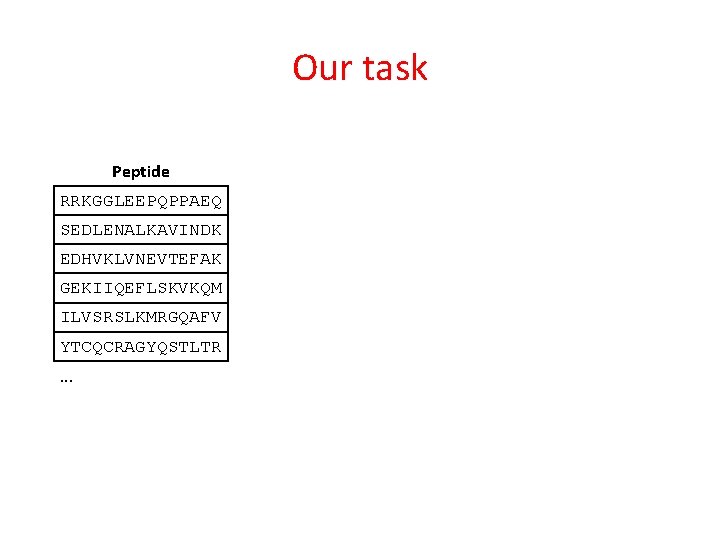
Our task Peptide RRKGGLEEPQPPAEQ SEDLENALKAVINDK EDHVKLVNEVTEFAK GEKIIQEFLSKVKQM ILVSRSLKMRGQAFV YTCQCRAGYQSTLTR. . .

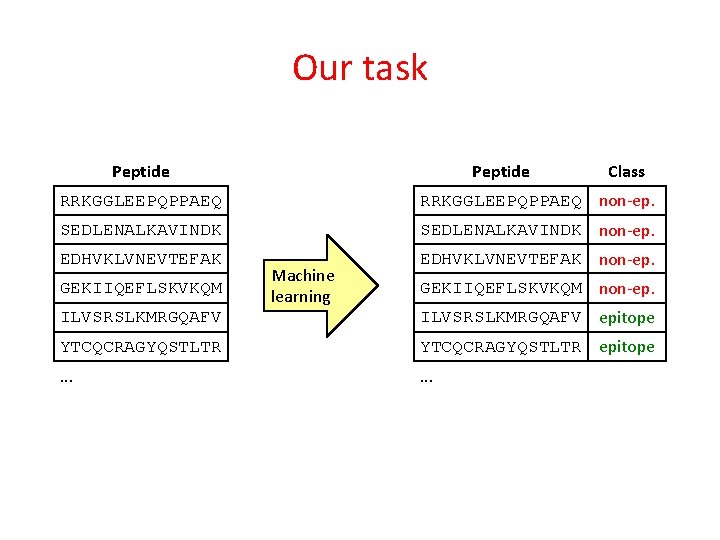
Our task Peptide Class RRKGGLEEPQPPAEQ non-ep. SEDLENALKAVINDK non-ep. EDHVKLVNEVTEFAK non-ep. GEKIIQEFLSKVKQM Machine learning GEKIIQEFLSKVKQM non-ep. ILVSRSLKMRGQAFV epitope YTCQCRAGYQSTLTR epitope . . .

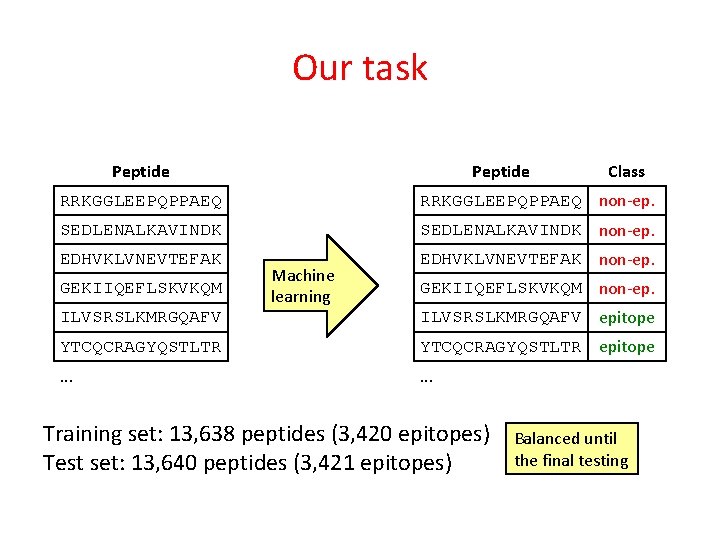
Our task Peptide Class RRKGGLEEPQPPAEQ non-ep. SEDLENALKAVINDK non-ep. EDHVKLVNEVTEFAK non-ep. GEKIIQEFLSKVKQM Machine learning GEKIIQEFLSKVKQM non-ep. ILVSRSLKMRGQAFV epitope YTCQCRAGYQSTLTR epitope . . . Training set: 13, 638 peptides (3, 420 epitopes) Test set: 13, 640 peptides (3, 421 epitopes) Balanced until the final testing

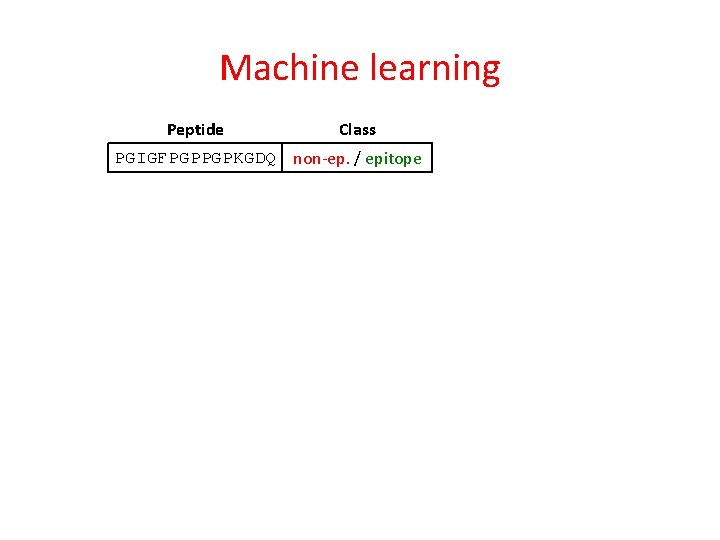
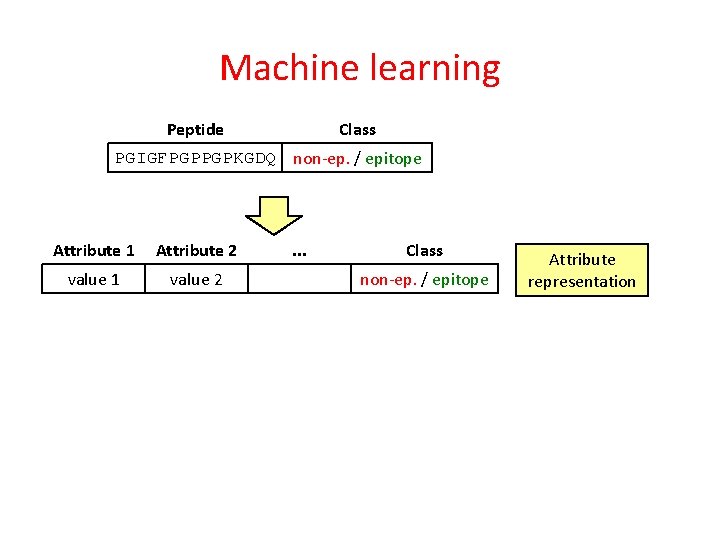
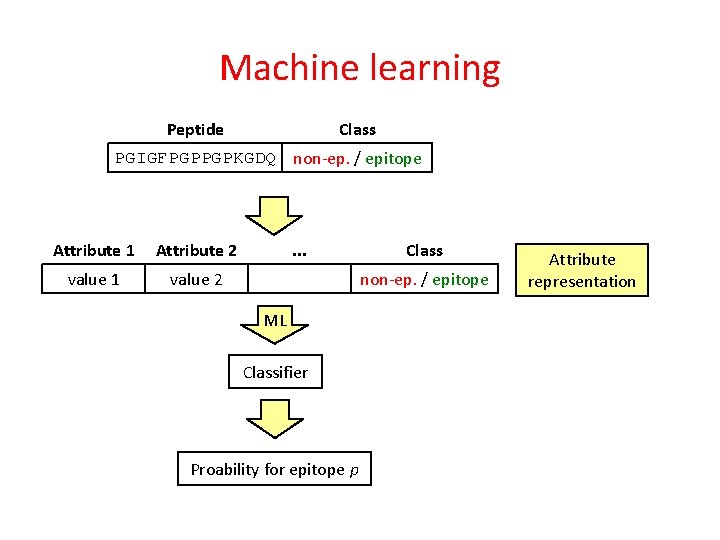
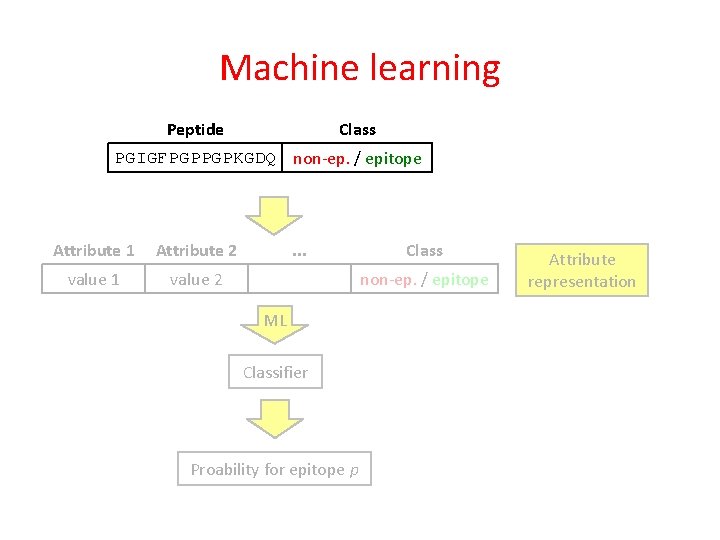
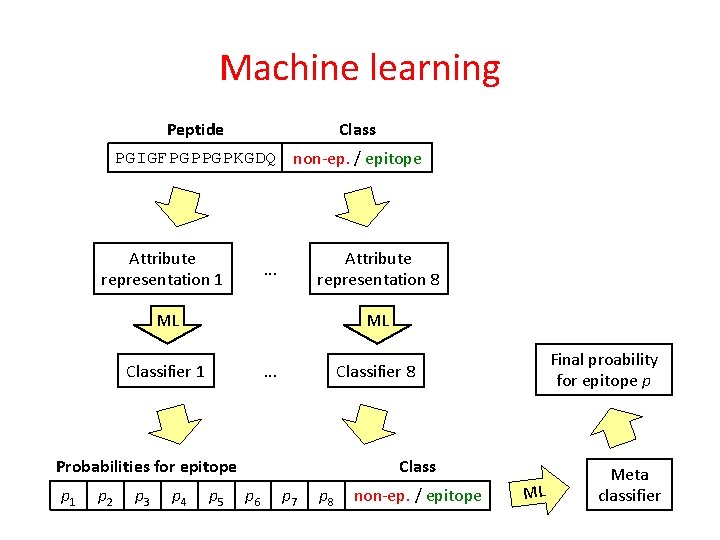
Machine learning Peptide Class PGIGFPGPPGPKGDQ non-ep. / epitope

Machine learning Class Peptide PGIGFPGPPGPKGDQ non-ep. / epitope Attribute 1 Attribute 2 value 1 value 2 . . . Class non-ep. / epitope Attribute representation

Machine learning Class Peptide PGIGFPGPPGPKGDQ non-ep. / epitope Attribute 1 Attribute 2 value 1 value 2 . . . Class non-ep. / epitope ML Classifier Proability for epitope p Attribute representation

Machine learning Class Peptide PGIGFPGPPGPKGDQ non-ep. / epitope Attribute 1 Attribute 2 value 1 value 2 . . . Class non-ep. / epitope ML Classifier Proability for epitope p Attribute representation

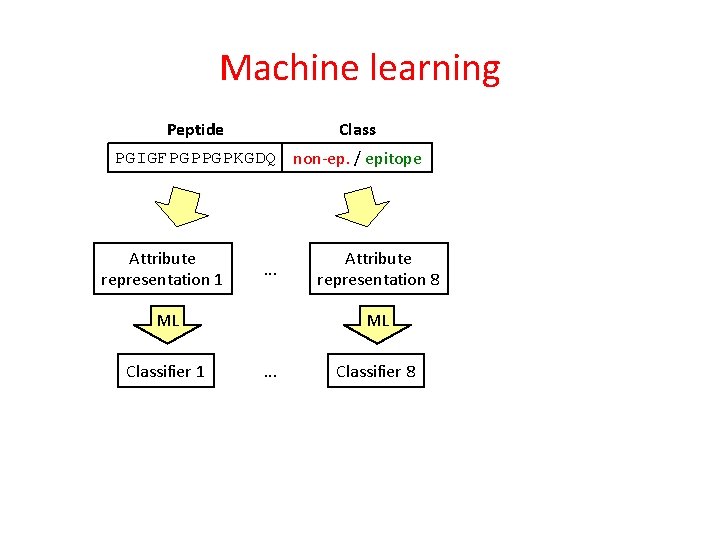
Machine learning Class Peptide PGIGFPGPPGPKGDQ non-ep. / epitope Attribute representation 1 . . . ML Classifier 1 Attribute representation 8 ML . . . Classifier 8

Machine learning Class Peptide PGIGFPGPPGPKGDQ non-ep. / epitope Attribute representation 1 Attribute representation 8 . . . ML ML Classifier 1 . . . Probabilities for epitope p 1 p 2 p 3 p 4 p 5 Final proability for epitope p Classifier 8 Class p 6 p 7 p 8 non-ep. / epitope ML Meta classifier

Machine learning Class Peptide PGIGFPGPPGPKGDQ non-ep. / epitope Attribute representation 1 SVM (SMO), Logistic regression Attribute representation 8 . . . ML ML Classifier 1 . . . Final proability for epitope p Classifier 8 Linear regression Probabilities for epitope p 1 p 2 p 3 p 4 p 5 Class p 6 p 7 p 8 non-ep. / epitope ML Meta classifier

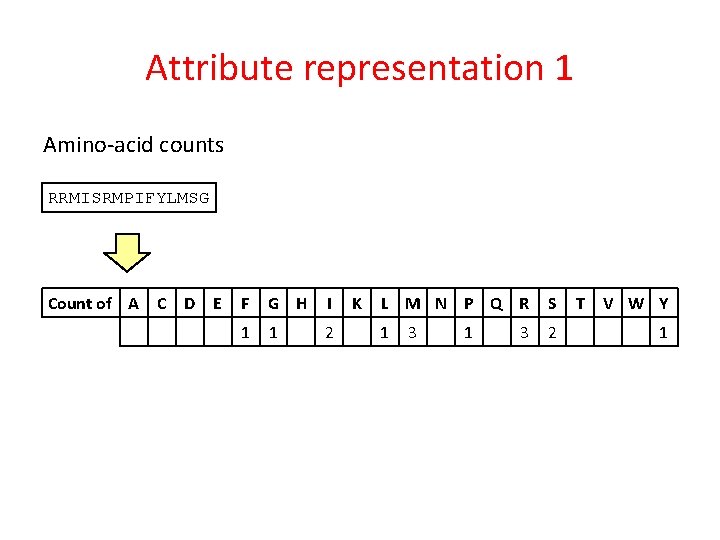
Attribute representation 1 Amino-acid counts RRMISRMPIFYLMSG Count of A C D E F G H I 1 1 2 K L M N P Q R S 1 2 3 1 3 T V W Y 1

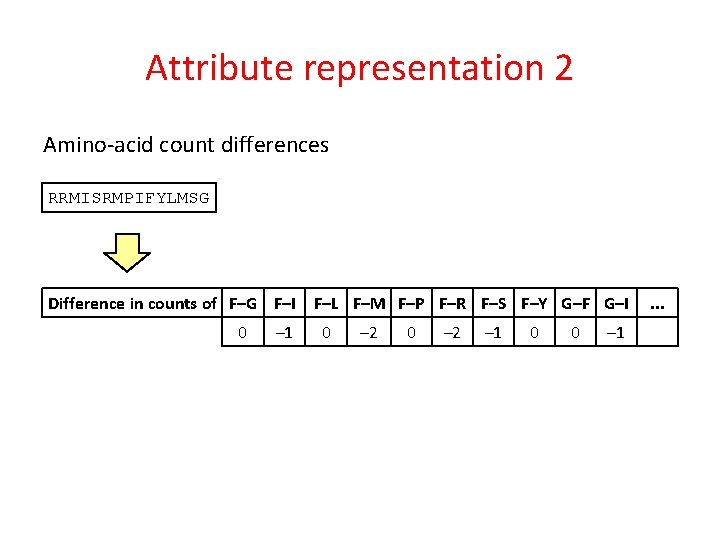
Attribute representation 2 Amino-acid count differences RRMISRMPIFYLMSG Difference in counts of F–G F–I 0 – 1 F–L F–M F–P F–R F–S F–Y G–F G–I 0 – 2 – 1 0 0 – 1 . . .

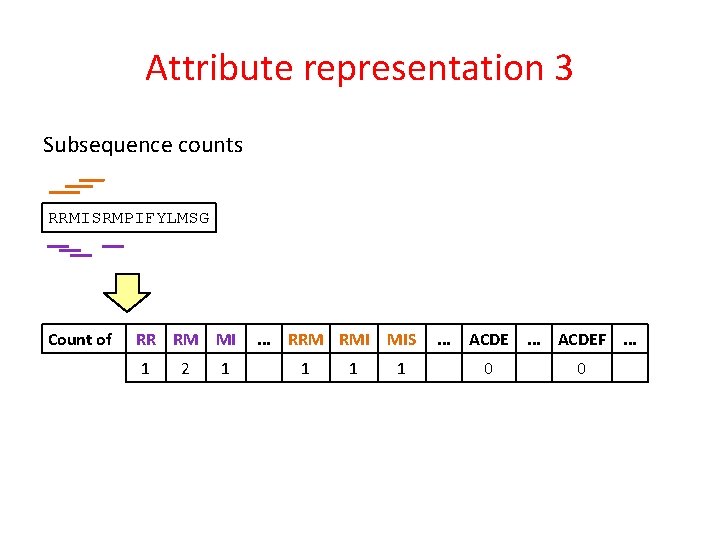
Attribute representation 3 Subsequence counts RRMISRMPIFYLMSG Count of RR RM MI 1 2 1 . . . RRM RMI 1 1 MIS 1 . . . ACDEF. . . 0 0

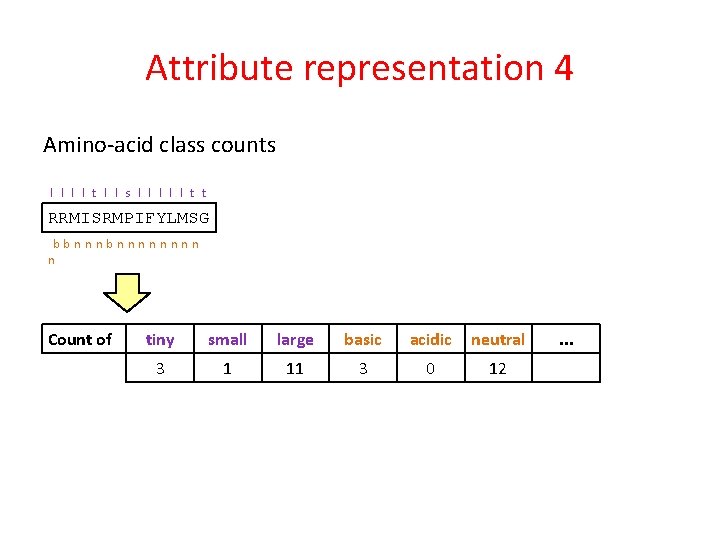
Attribute representation 4 Amino-acid class counts l l t l l s l l l t t RRMISRMPIFYLMSG bbnnnnnnnn n Count of tiny small large basic acidic neutral 3 1 11 3 0 12 . . .

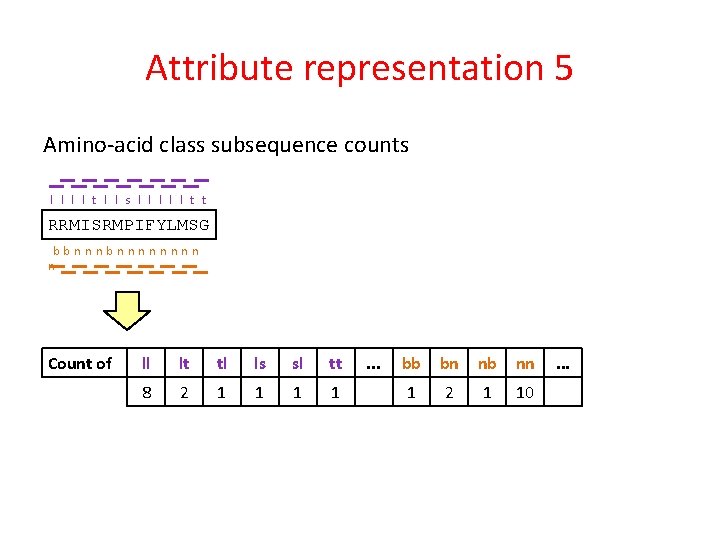
Attribute representation 5 Amino-acid class subsequence counts l l t l l s l l l t t RRMISRMPIFYLMSG bbnnnnnnnn n Count of ll lt tl ls sl tt 8 2 1 1 . . . bb bn nb nn 1 2 1 10 . . .

Attribute representation 6 Amino-acid pair counts Rationale: antibodies may bind in two places due to their twochain structure. Antibody Peptide

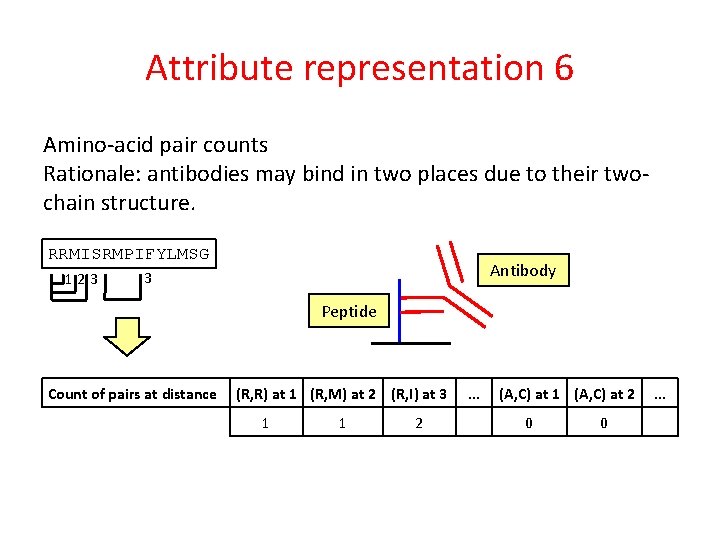
Attribute representation 6 Amino-acid pair counts Rationale: antibodies may bind in two places due to their twochain structure. RRMISRMPIFYLMSG 123 Antibody 3 Peptide Count of pairs at distance (R, R) at 1 (R, M) at 2 (R, I) at 3 1 1 2 . . . (A, C) at 1 (A, C) at 2 0 0 . . .

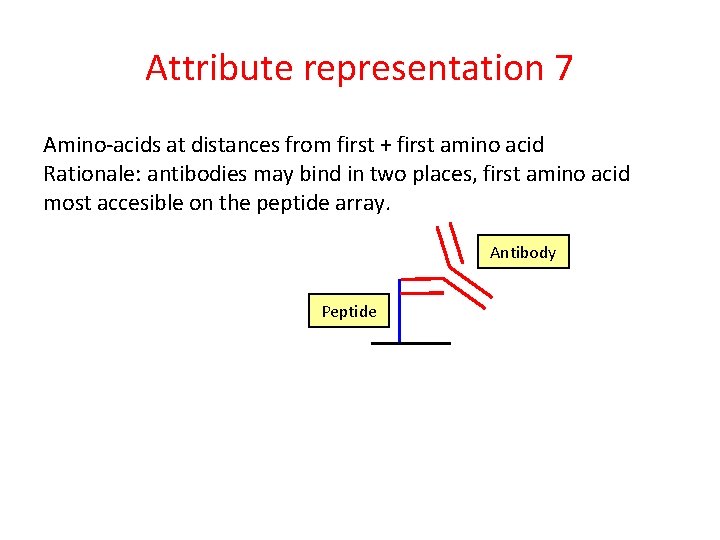
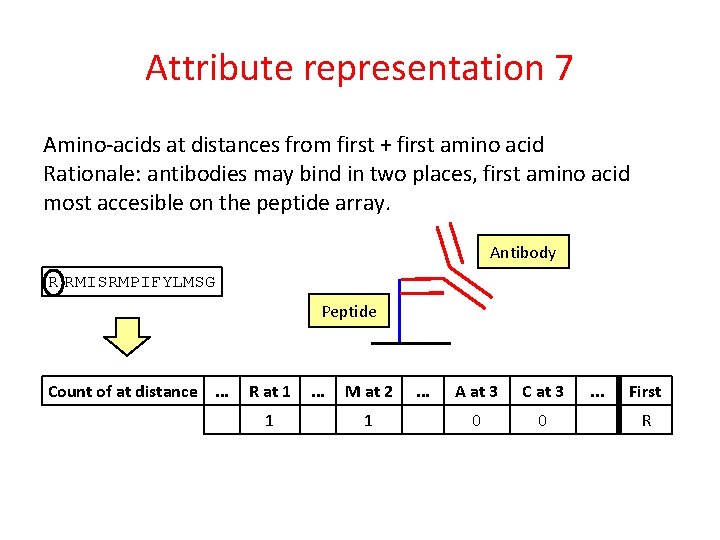
Attribute representation 7 Amino-acids at distances from first + first amino acid Rationale: antibodies may bind in two places, first amino acid most accesible on the peptide array. Antibody Peptide

Attribute representation 7 Amino-acids at distances from first + first amino acid Rationale: antibodies may bind in two places, first amino acid most accesible on the peptide array. Antibody R RMISRMPIFYLMSG Peptide Count of at distance. . . R at 1 1 . . . M at 2 1 . . . A at 3 C at 3 0 0 . . . First R

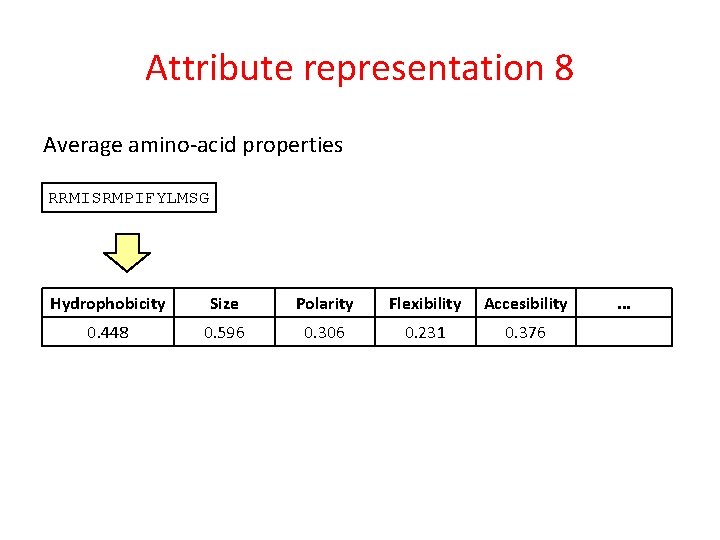
Attribute representation 8 Average amino-acid properties RRMISRMPIFYLMSG Hydrophobicity Size Polarity Flexibility Accesibility 0. 448 0. 596 0. 306 0. 231 0. 376 . . .

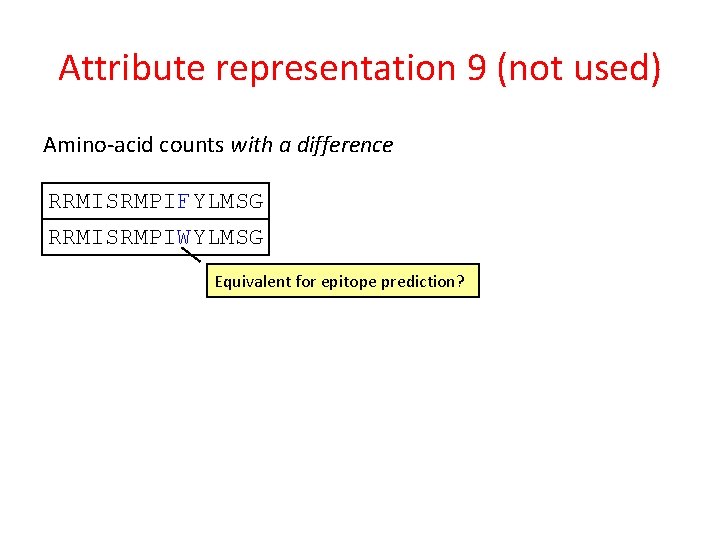
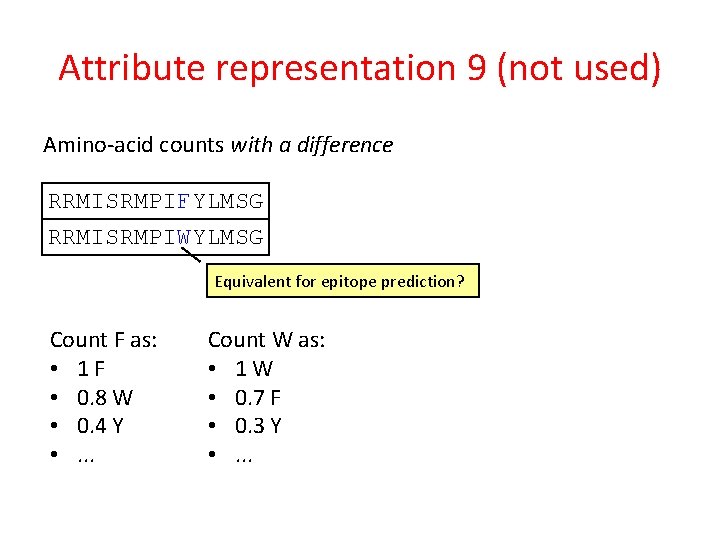
Attribute representation 9 (not used) Amino-acid counts with a difference RRMISRMPIFYLMSG RRMISRMPIWYLMSG Equivalent for epitope prediction?

Attribute representation 9 (not used) Amino-acid counts with a difference RRMISRMPIFYLMSG RRMISRMPIWYLMSG Equivalent for epitope prediction? Count F as: • 1 F • 0. 8 W • 0. 4 Y • . . . Count W as: • 1 W • 0. 7 F • 0. 3 Y • . . .

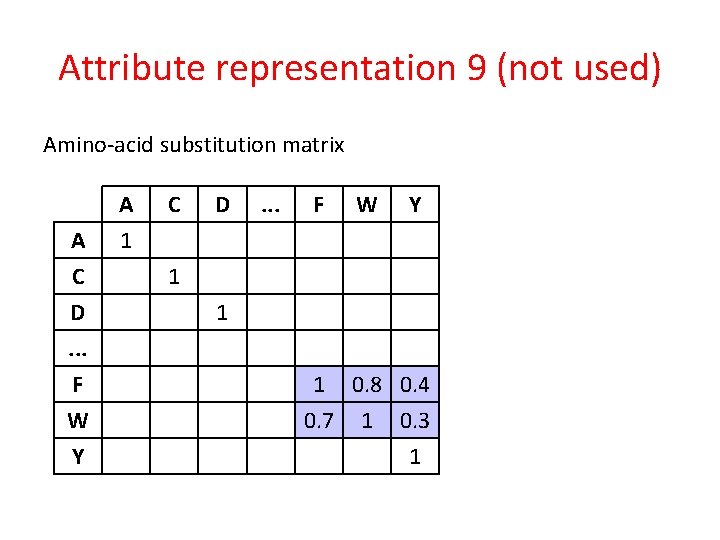
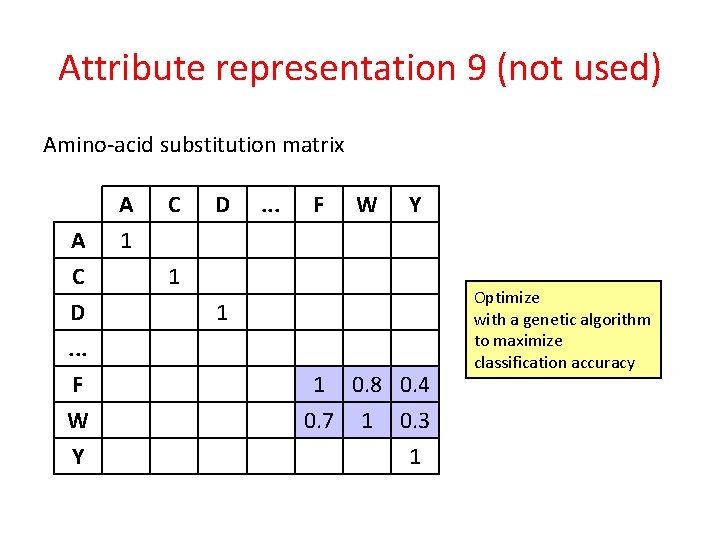
Attribute representation 9 (not used) Amino-acid substitution matrix A C D. . . F W Y A 1 C D . . . F W Y 1 1 1 0. 8 0. 4 0. 7 1 0. 3 1

Attribute representation 9 (not used) Amino-acid substitution matrix A C D. . . F W Y A 1 C D . . . F W Y 1 1 1 0. 8 0. 4 0. 7 1 0. 3 1 Optimize with a genetic algorithm to maximize classification accuracy

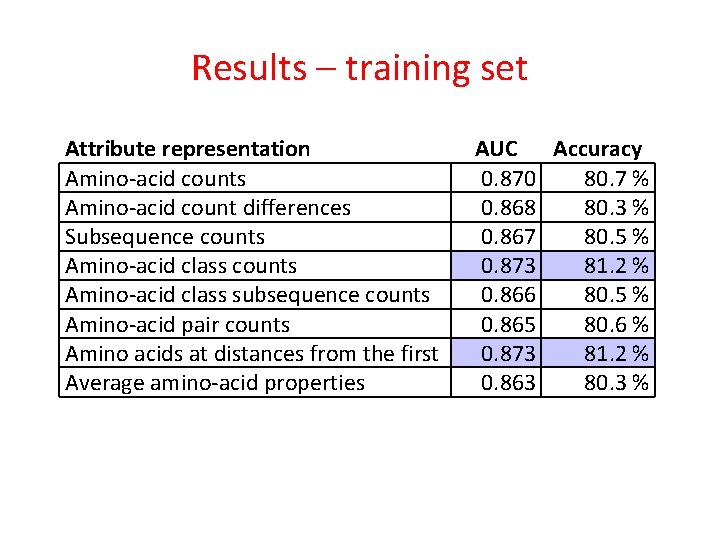
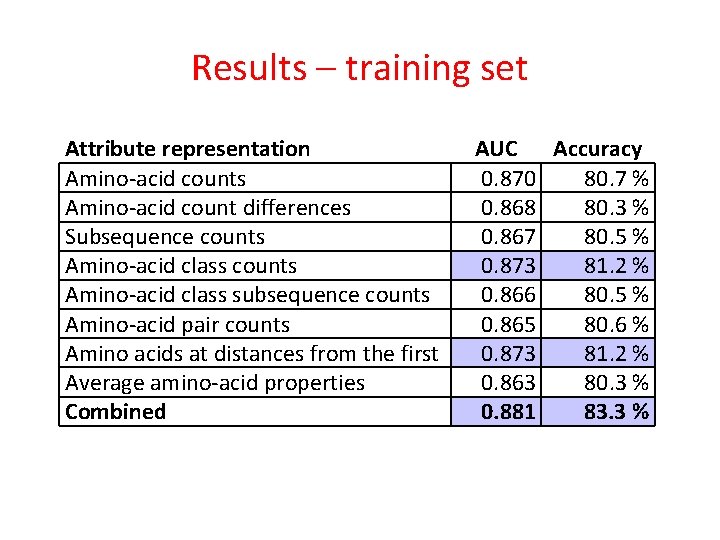
Results – training set Attribute representation Amino-acid counts Amino-acid count differences Subsequence counts Amino-acid class subsequence counts Amino-acid pair counts Amino acids at distances from the first Average amino-acid properties AUC Accuracy 0. 870 80. 7 % 0. 868 80. 3 % 0. 867 80. 5 % 0. 873 81. 2 % 0. 866 80. 5 % 0. 865 80. 6 % 0. 873 81. 2 % 0. 863 80. 3 %

Results – training set Attribute representation Amino-acid counts Amino-acid count differences Subsequence counts Amino-acid class subsequence counts Amino-acid pair counts Amino acids at distances from the first Average amino-acid properties Combined AUC Accuracy 0. 870 80. 7 % 0. 868 80. 3 % 0. 867 80. 5 % 0. 873 81. 2 % 0. 866 80. 5 % 0. 865 80. 6 % 0. 873 81. 2 % 0. 863 80. 3 % 0. 881 83. 3 %

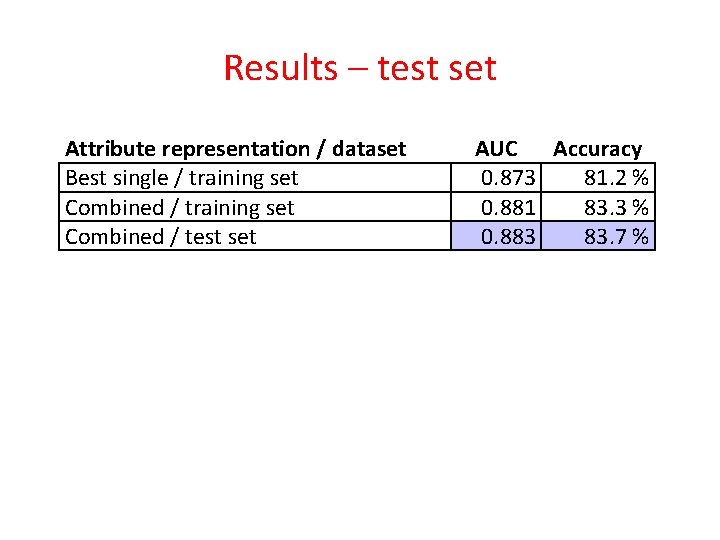
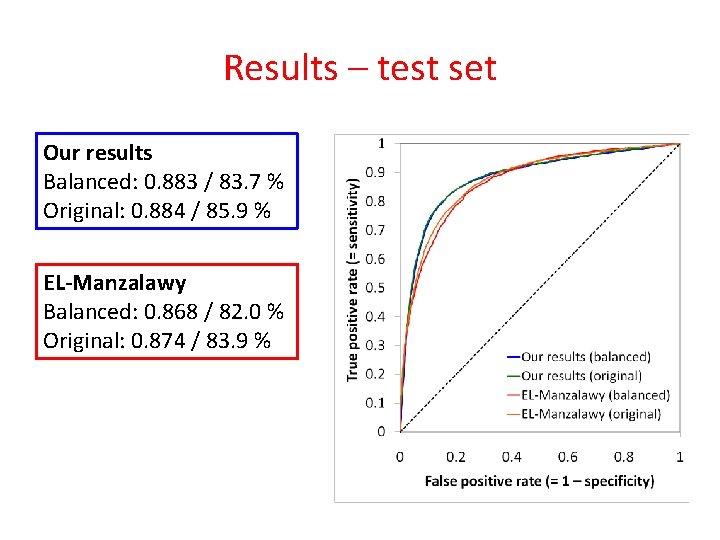
Results – test set Attribute representation / dataset Best single / training set Combined / test set AUC Accuracy 0. 873 81. 2 % 0. 881 83. 3 % 0. 883 83. 7 %

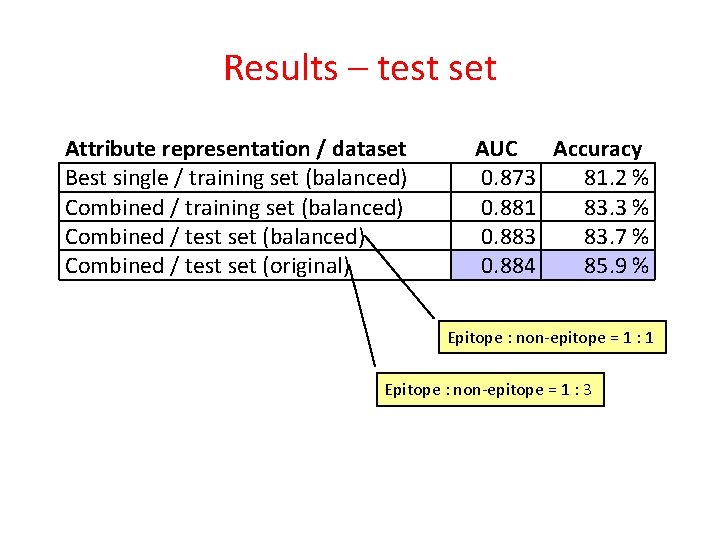
Results – test set Attribute representation / dataset Best single / training set (balanced) Combined / test set (original) AUC Accuracy 0. 873 81. 2 % 0. 881 83. 3 % 0. 883 83. 7 % 0. 884 85. 9 % Epitope : non-epitope = 1 : 1 Epitope : non-epitope = 1 : 3

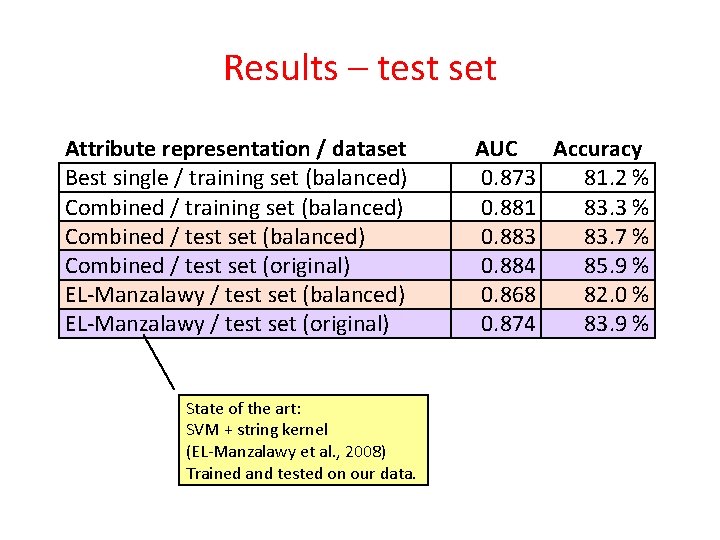
Results – test set Attribute representation / dataset Best single / training set (balanced) Combined / test set (original) EL-Manzalawy / test set (balanced) EL-Manzalawy / test set (original) State of the art: SVM + string kernel (EL-Manzalawy et al. , 2008) Trained and tested on our data. AUC Accuracy 0. 873 81. 2 % 0. 881 83. 3 % 0. 883 83. 7 % 0. 884 85. 9 % 0. 868 82. 0 % 0. 874 83. 9 %

Results – test set Our results Balanced: 0. 883 / 83. 7 % Original: 0. 884 / 85. 9 % EL-Manzalawy Balanced: 0. 868 / 82. 0 % Original: 0. 874 / 83. 9 %

1. Introduction 2. Immune response prediction 3. Interpretation

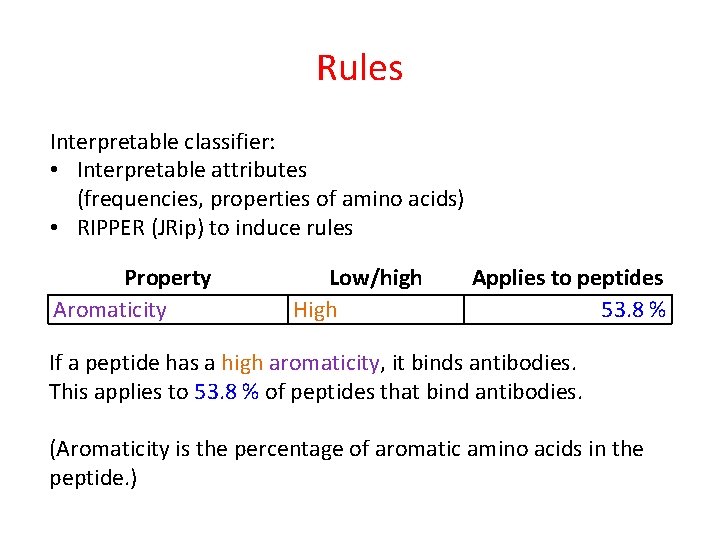
Rules Interpretable classifier: • Interpretable attributes (frequencies, properties of amino acids) • RIPPER (JRip) to induce rules

Rules Interpretable classifier: • Interpretable attributes (frequencies, properties of amino acids) • RIPPER (JRip) to induce rules Property Aromaticity Low/high High Applies to peptides 53. 8 % If a peptide has a high aromaticity, it binds antibodies. This applies to 53. 8 % of peptides that bind antibodies. (Aromaticity is the percentage of aromatic amino acids in the peptide. )

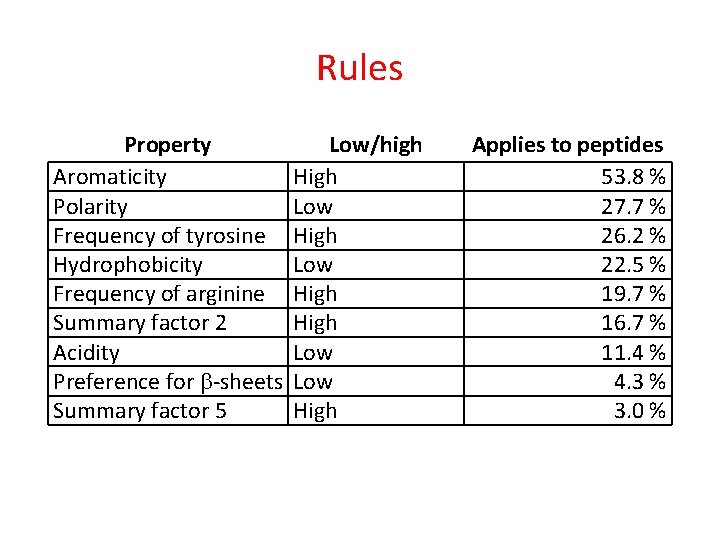
Rules Property Aromaticity Polarity Frequency of tyrosine Hydrophobicity Frequency of arginine Summary factor 2 Acidity Preference for -sheets Summary factor 5 Low/high High Low High Applies to peptides 53. 8 % 27. 7 % 26. 2 % 22. 5 % 19. 7 % 16. 7 % 11. 4 % 4. 3 % 3. 0 %

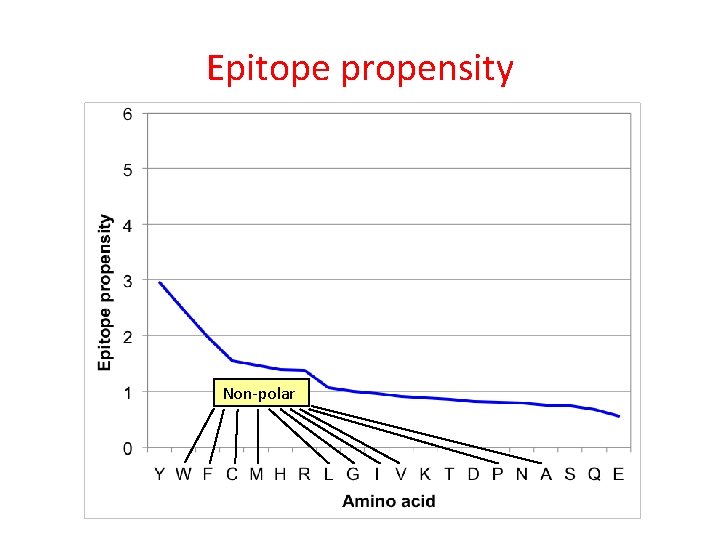
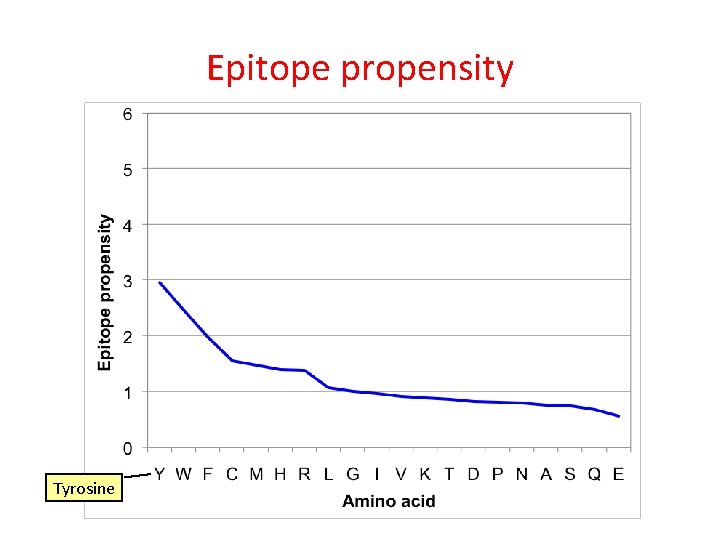
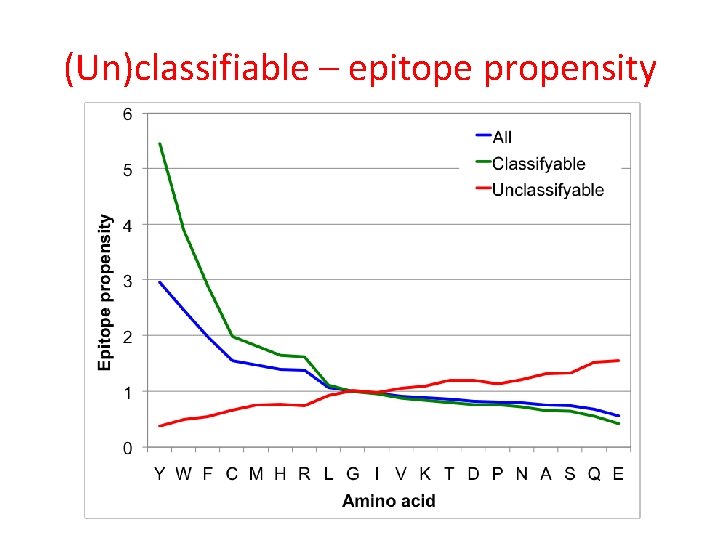
Epitope propensity Frequency in peptides with epitopes, divided by frequency in peptides without epitopes

Epitope propensity Aromatic

Epitope propensity Non-polar

Epitope propensity Tyrosine

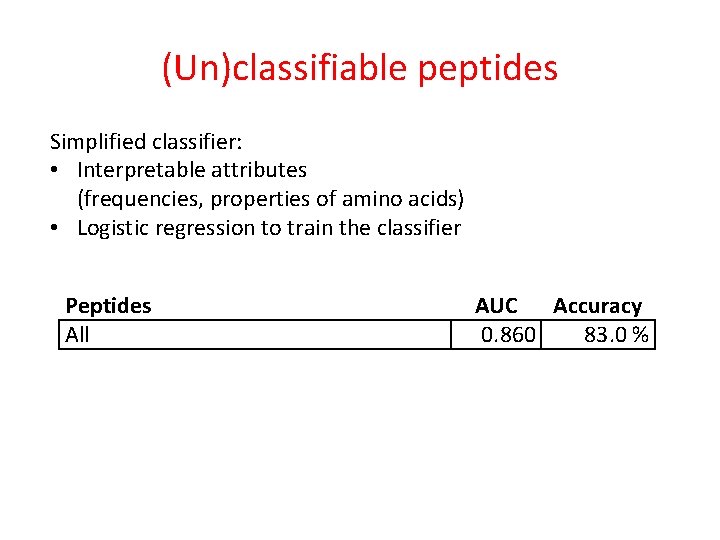
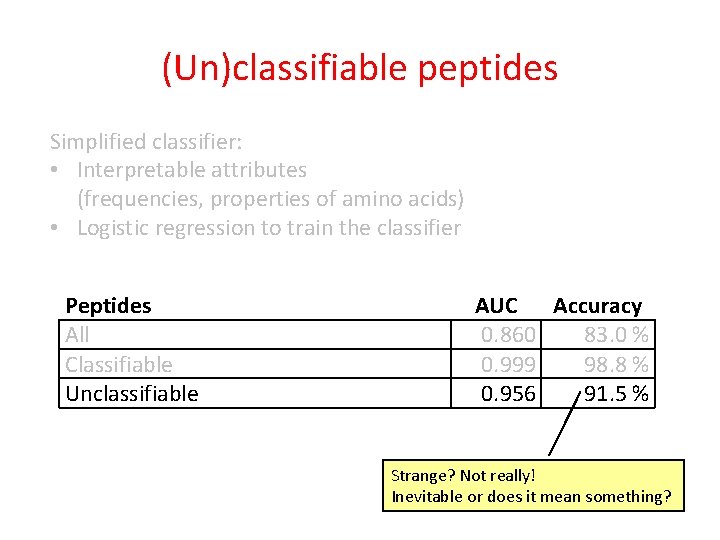
(Un)classifiable peptides Simplified classifier: • Interpretable attributes (frequencies, properties of amino acids) • Logistic regression to train the classifier Peptides All AUC Accuracy 0. 860 83. 0 %

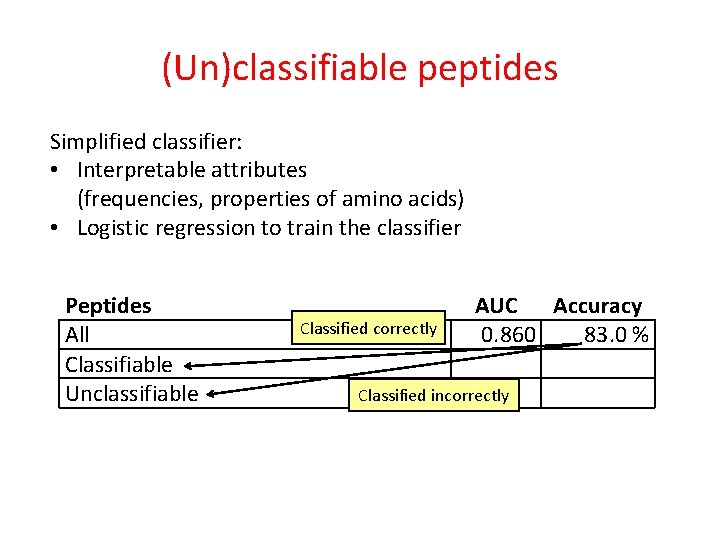
(Un)classifiable peptides Simplified classifier: • Interpretable attributes (frequencies, properties of amino acids) • Logistic regression to train the classifier Peptides All Classifiable Unclassifiable Classified correctly AUC Accuracy 0. 860 83. 0 % Classified incorrectly

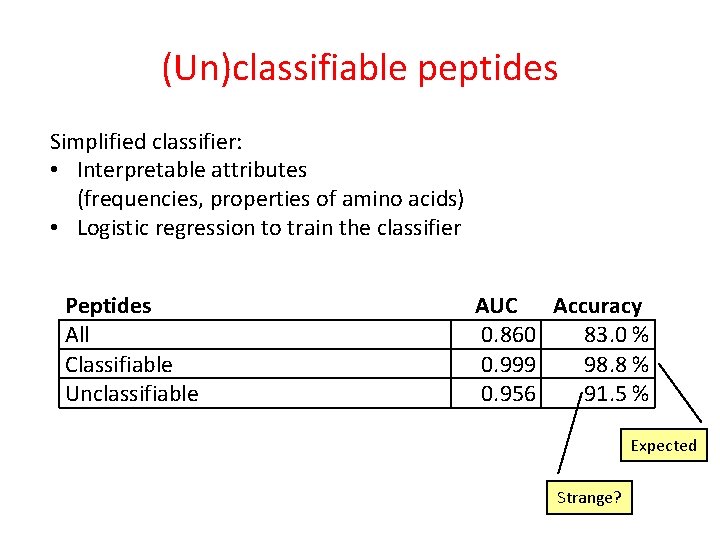
(Un)classifiable peptides Simplified classifier: • Interpretable attributes (frequencies, properties of amino acids) • Logistic regression to train the classifier Peptides All Classifiable Unclassifiable AUC Accuracy 0. 860 83. 0 % 0. 999 98. 8 % 0. 956 91. 5 % Expected Strange?

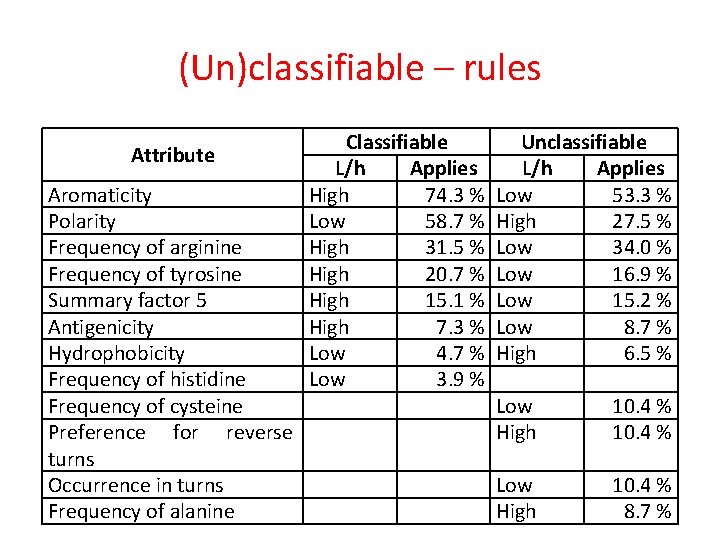
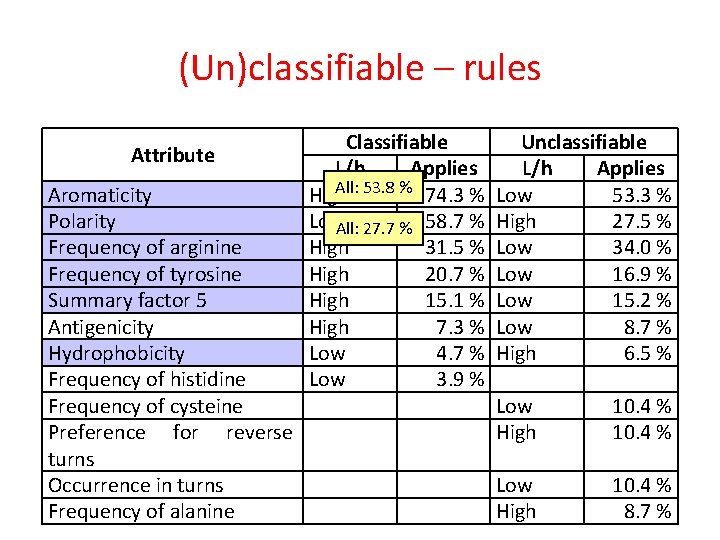
(Un)classifiable – rules Attribute Aromaticity Polarity Frequency of arginine Frequency of tyrosine Summary factor 5 Antigenicity Hydrophobicity Frequency of histidine Frequency of cysteine Preference for reverse turns Occurrence in turns Frequency of alanine Classifiable L/h Applies High 74. 3 % Low 58. 7 % High 31. 5 % High 20. 7 % High 15. 1 % High 7. 3 % Low 4. 7 % Low 3. 9 % Unclassifiable L/h Applies Low 53. 3 % High 27. 5 % Low 34. 0 % Low 16. 9 % Low 15. 2 % Low 8. 7 % High 6. 5 % Low High 10. 4 % 8. 7 %

(Un)classifiable – rules Attribute Aromaticity Polarity Frequency of arginine Frequency of tyrosine Summary factor 5 Antigenicity Hydrophobicity Frequency of histidine Frequency of cysteine Preference for reverse turns Occurrence in turns Frequency of alanine Classifiable L/h Applies All: 53. 8 % 74. 3 % High Low. All: 27. 7 % 58. 7 % High 31. 5 % High 20. 7 % High 15. 1 % High 7. 3 % Low 4. 7 % Low 3. 9 % Unclassifiable L/h Applies Low 53. 3 % High 27. 5 % Low 34. 0 % Low 16. 9 % Low 15. 2 % Low 8. 7 % High 6. 5 % Low High 10. 4 % 8. 7 %

(Un)classifiable – epitope propensity

(Un)classifiable peptides Simplified classifier: • Interpretable attributes (frequencies, properties of amino acids) • Logistic regression to train the classifier Peptides All Classifiable Unclassifiable AUC Accuracy 0. 860 83. 0 % 0. 999 98. 8 % 0. 956 91. 5 % Strange? Not really! Inevitable or does it mean something?

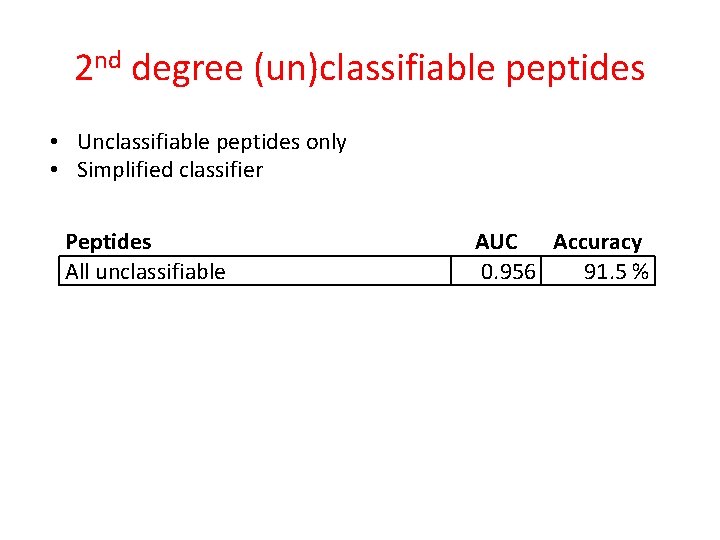
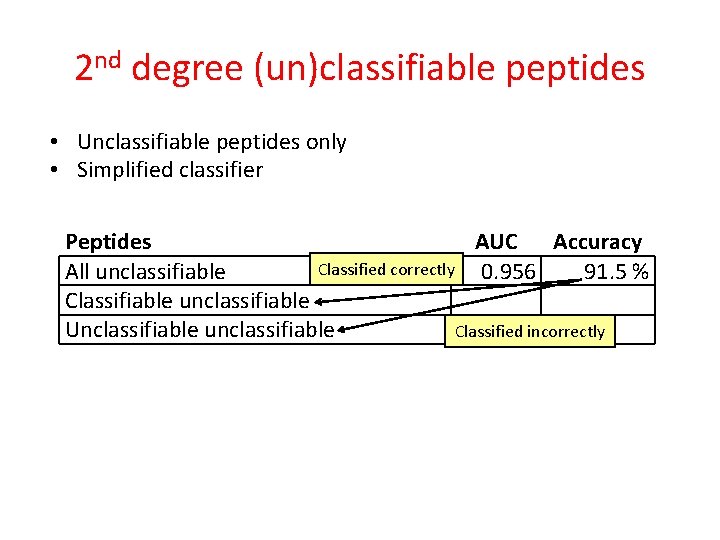
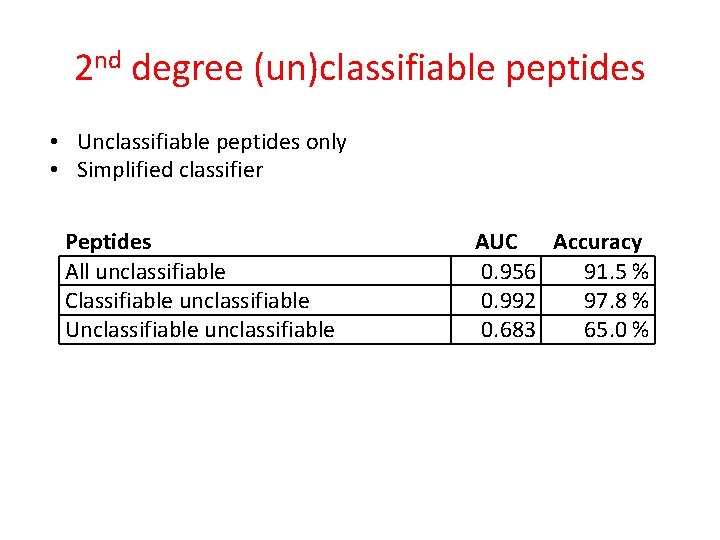
2 nd degree (un)classifiable peptides • Unclassifiable peptides only • Simplified classifier Peptides All unclassifiable AUC Accuracy 0. 956 91. 5 %

2 nd degree (un)classifiable peptides • Unclassifiable peptides only • Simplified classifier Peptides AUC Accuracy Classified correctly 0. 956 All unclassifiable 91. 5 % Classifiable unclassifiable Classified incorrectly Unclassifiable unclassifiable

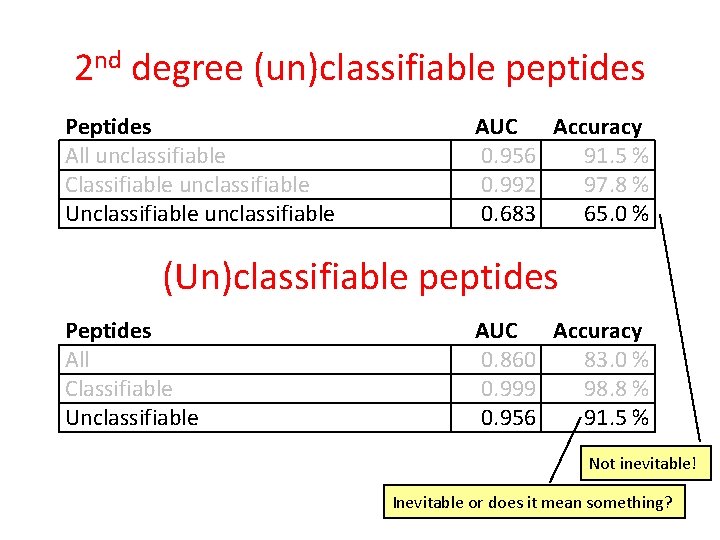
2 nd degree (un)classifiable peptides • Unclassifiable peptides only • Simplified classifier Peptides All unclassifiable Classifiable unclassifiable Unclassifiable unclassifiable AUC Accuracy 0. 956 91. 5 % 0. 992 97. 8 % 0. 683 65. 0 %

2 nd degree (un)classifiable peptides Peptides All unclassifiable Classifiable unclassifiable Unclassifiable unclassifiable AUC Accuracy 0. 956 91. 5 % 0. 992 97. 8 % 0. 683 65. 0 % (Un)classifiable peptides Peptides All Classifiable Unclassifiable AUC Accuracy 0. 860 83. 0 % 0. 999 98. 8 % 0. 956 91. 5 % Not inevitable! Inevitable or does it mean something?

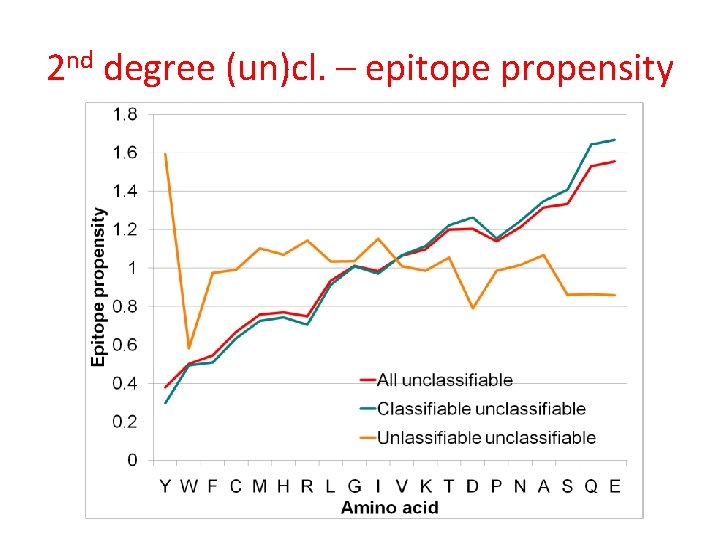
2 nd degree (un)cl. – epitope propensity

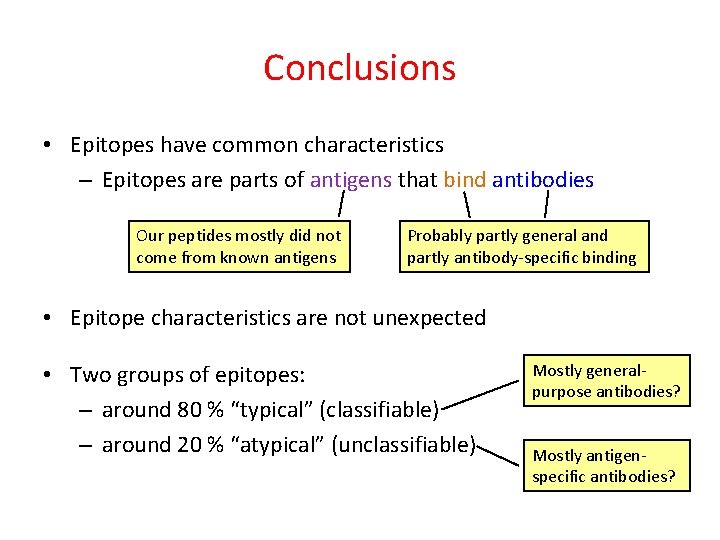
Conclusions • Epitopes have common characteristics

Conclusions • Epitopes have common characteristics – Epitopes are parts of antigens that bind antibodies Our peptides mostly did not come from known antigens Probably partly general and partly antibody-specific binding

Conclusions • Epitopes have common characteristics – Epitopes are parts of antigens that bind antibodies Our peptides mostly did not come from known antigens Probably partly general and partly antibody-specific binding • Epitope characteristics are not unexpected

Conclusions • Epitopes have common characteristics – Epitopes are parts of antigens that bind antibodies Our peptides mostly did not come from known antigens Probably partly general and partly antibody-specific binding • Epitope characteristics are not unexpected • Two groups of epitopes: – around 80 % “typical” (classifiable) – around 20 % “atypical” (unclassifiable)

Conclusions • Epitopes have common characteristics – Epitopes are parts of antigens that bind antibodies Our peptides mostly did not come from known antigens Probably partly general and partly antibody-specific binding • Epitope characteristics are not unexpected • Two groups of epitopes: – around 80 % “typical” (classifiable) – around 20 % “atypical” (unclassifiable) Mostly generalpurpose antibodies? Mostly antigenspecific antibodies?




























