Marker asisted selection MAS in increasing production traits







- Slides: 31

Marker asisted selection (MAS) in increasing production traits of Friesian Holstein dairy cattle in Indonesia SUTARNO Biology Department, Faculty of Mathematics and Natural Sciences, Sebelas Maret University, Jl. Ir. Sutami 36 A Surakarta, 57126 Indonesia

Friesian Holstein Dairy Cattle • The first Indonesian FH cattle was imported from Australia, New Zealand USA, and reared in Java highland at 700 m above sea level • Good performance in reproduction and milk production • Able to adapt with indonesian climate under temperatures range 5 -250 C

Breeding Selection is one of the effort to improve genetic potential in animal population DNA based (Marker assisted selection ) Conventional (phenotype)

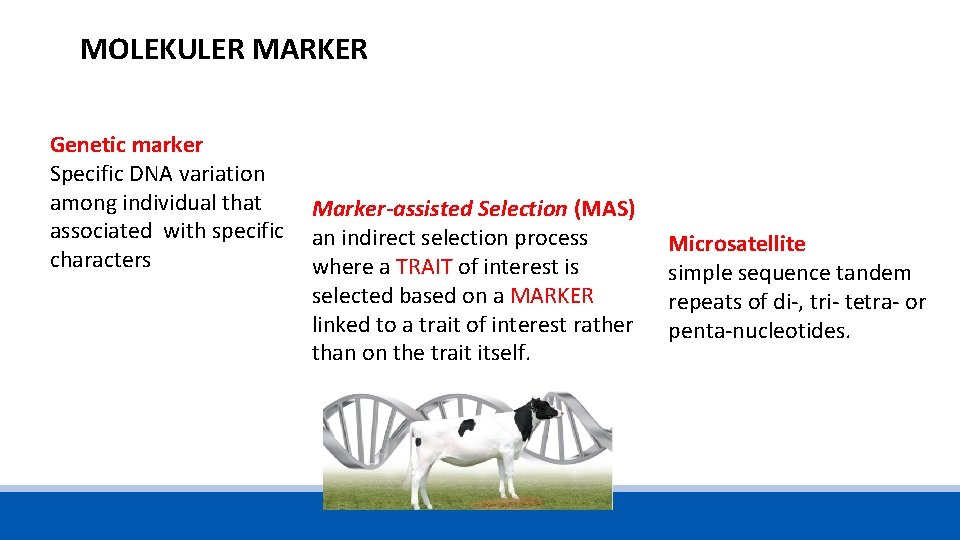
MOLEKULER MARKER Genetic marker Specific DNA variation among individual that associated with specific characters Marker-assisted Selection (MAS) an indirect selection process where a TRAIT of interest is selected based on a MARKER linked to a trait of interest rather than on the trait itself. Microsatellite simple sequence tandem repeats of di-, tri- tetra- or penta-nucleotides.

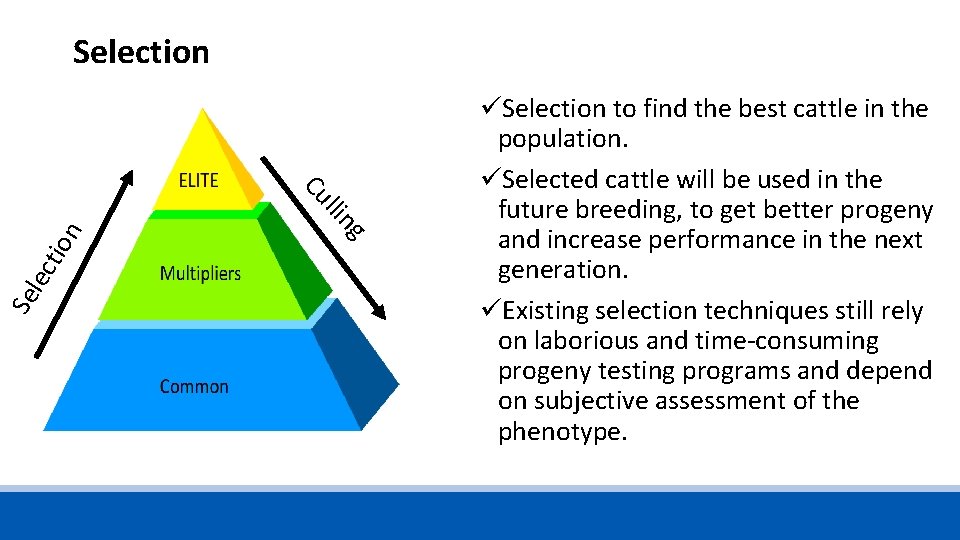
Selection n tio lec Se g llin Cu üSelection to find the best cattle in the population. üSelected cattle will be used in the future breeding, to get better progeny and increase performance in the next generation. üExisting selection techniques still rely on laborious and time-consuming progeny testing programs and depend on subjective assessment of the phenotype.

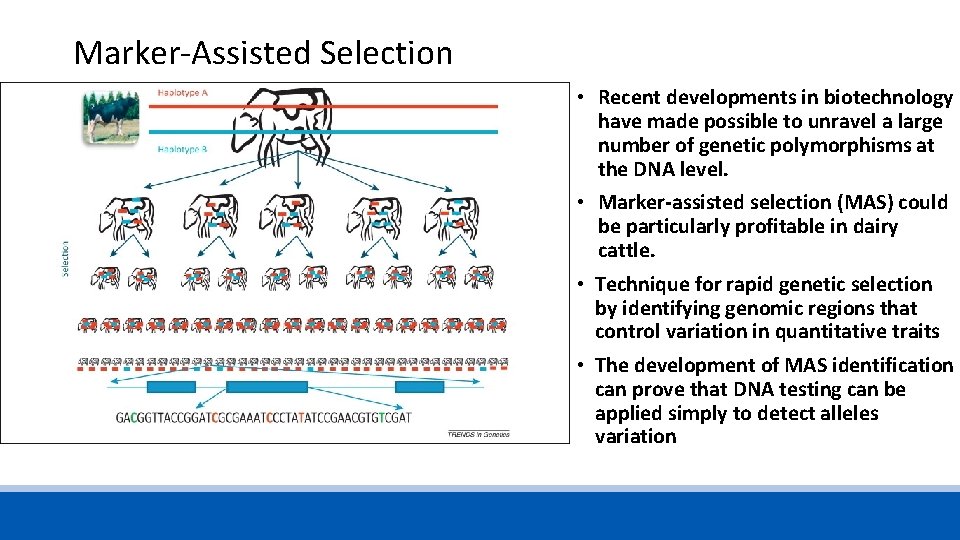
Marker-Assisted Selection • Recent developments in biotechnology have made possible to unravel a large number of genetic polymorphisms at the DNA level. • Marker-assisted selection (MAS) could be particularly profitable in dairy cattle. • Technique for rapid genetic selection by identifying genomic regions that control variation in quantitative traits • The development of MAS identification can prove that DNA testing can be applied simply to detect alleles variation

Microsatellites • Microsatellite, a new class of genetic marker, is a technique based on length variation within tandem arrays of di-, tri-, or tetra nucleotide motifs. • There are several published paper tried associating microsatellite and quantitative performance trait in the dairy cattle population.

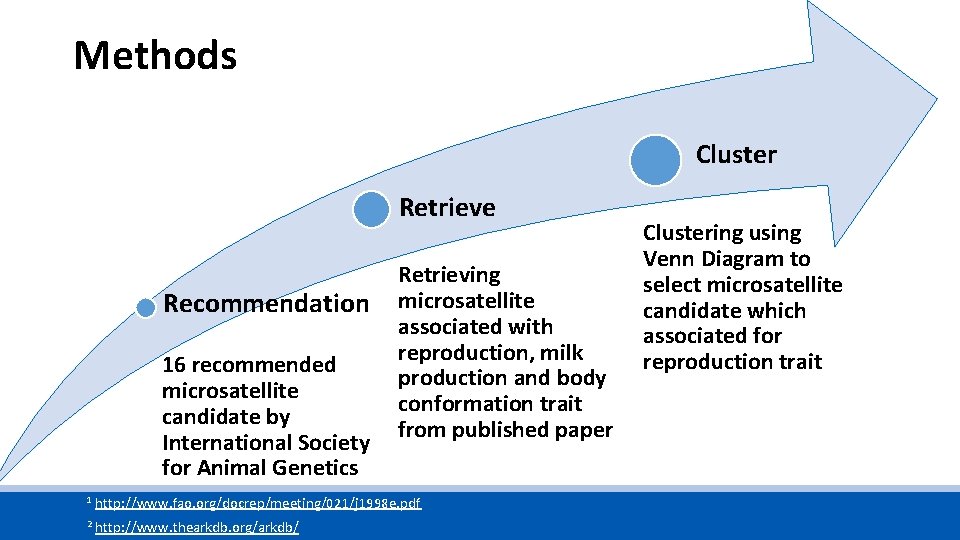
Methods Cluster Retrieve Recommendation 16 recommended microsatellite candidate by International Society for Animal Genetics Retrieving microsatellite associated with reproduction, milk production and body conformation trait from published paper 1 http: //www. fao. org/docrep/meeting/021/j 1998 e. pdf 2 http: //www. thearkdb. org/arkdb/ Clustering using Venn Diagram to select microsatellite candidate which associated for reproduction trait

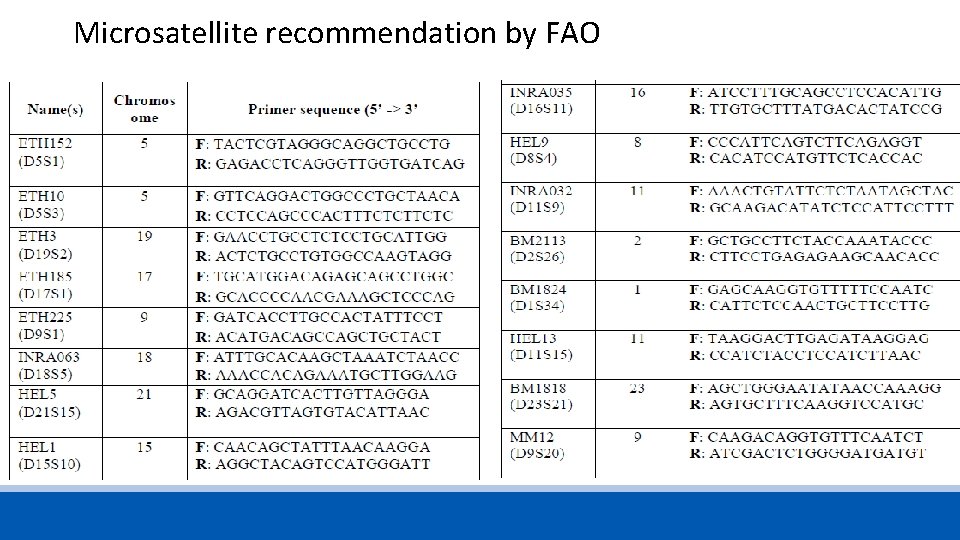
Microsatellite recommendation by FAO

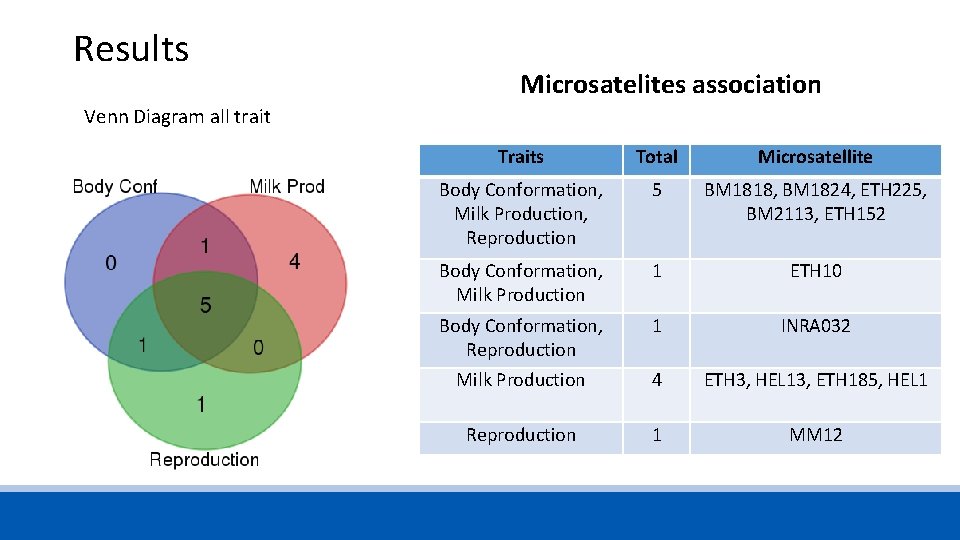
Results Microsatelites association Venn Diagram all trait Traits Total Microsatellite Body Conformation, Milk Production, Reproduction 5 BM 1818, BM 1824, ETH 225, BM 2113, ETH 152 Body Conformation, Milk Production 1 ETH 10 Body Conformation, Reproduction 1 INRA 032 Milk Production 4 ETH 3, HEL 13, ETH 185, HEL 1 Reproduction 1 MM 12

• Performance affected by interaction between genetic and environment factors (P = G x E) • Genotype-based selection for intersted phenotype traits.

Production/ performance traits of dairy cattle under study ØReproduction ØMilk Production ØBody Conformation

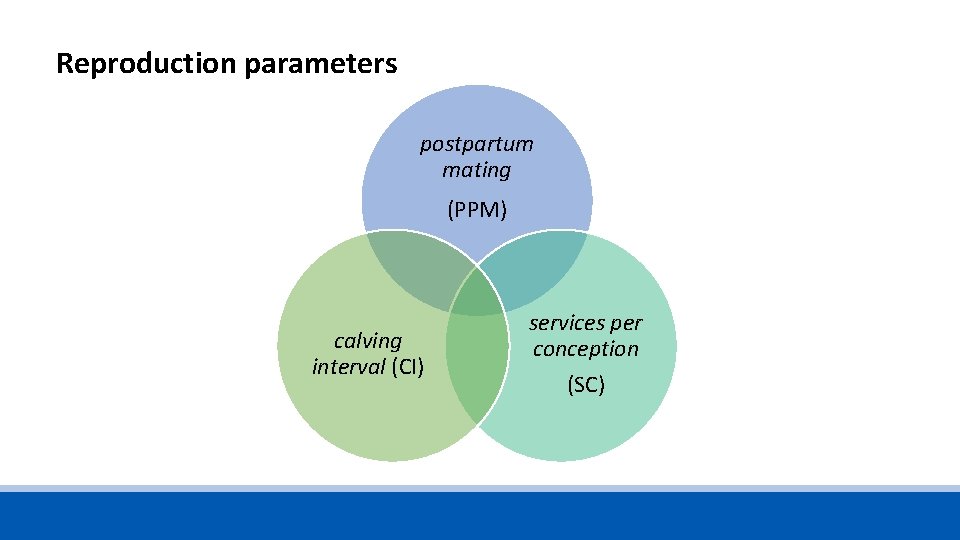
1. Reproduction • Cows start producing milk after partus and will ongoing if the reproduction process continues. • Milk performance and reproductive parameters are very important in term of management and profitability. • Reproductive performance is probably the most important factor that is a prerequisite for sustainable dairy production system and influencing the productivity.

Reproduction parameters postpartum mating (PPM) calving interval (CI) services per conception (SC)

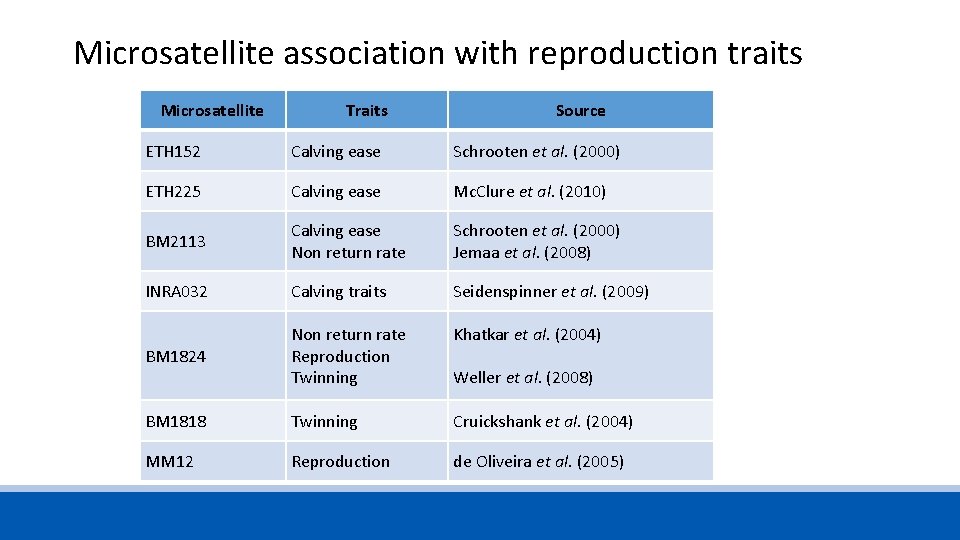
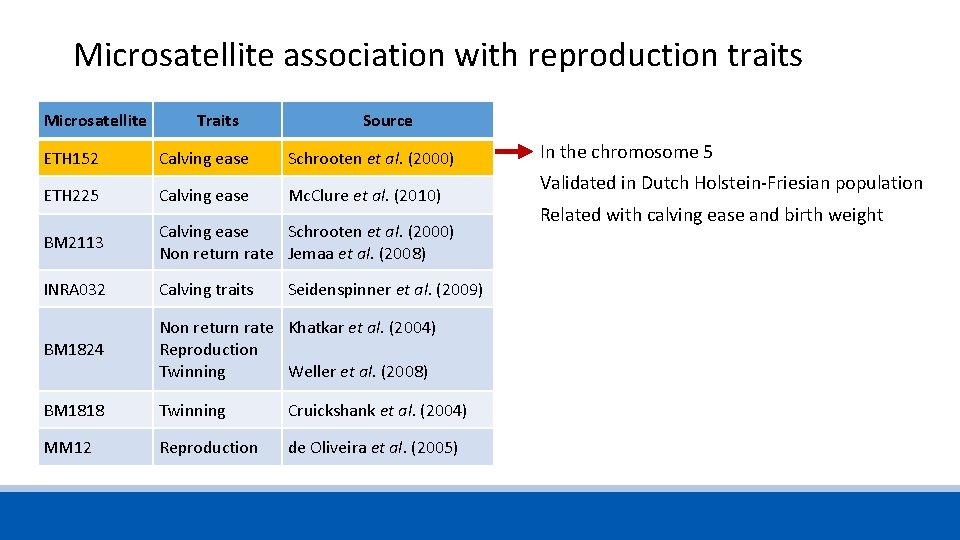
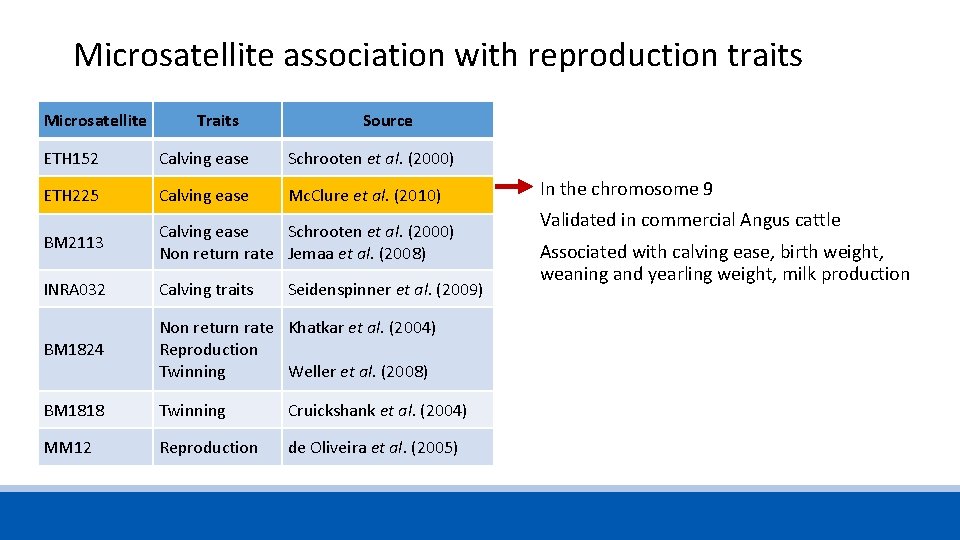
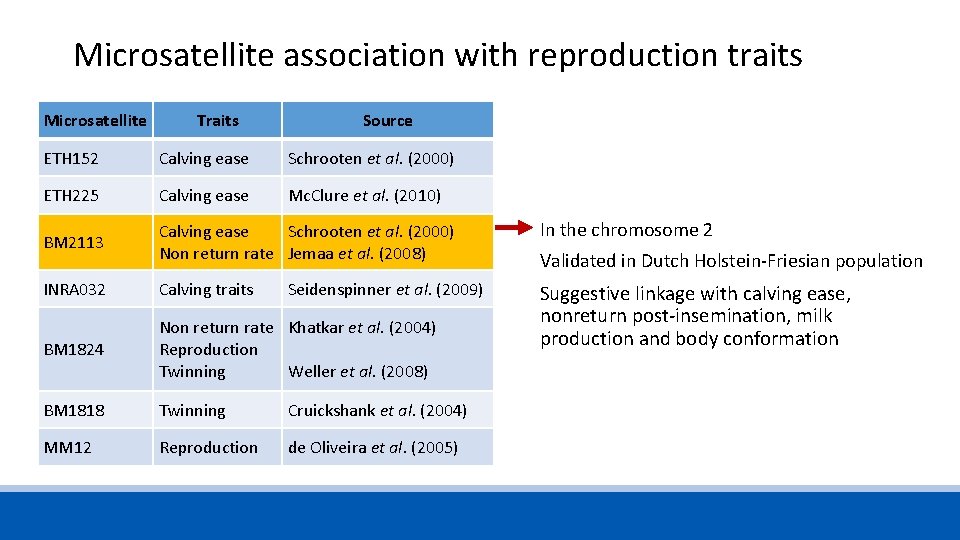
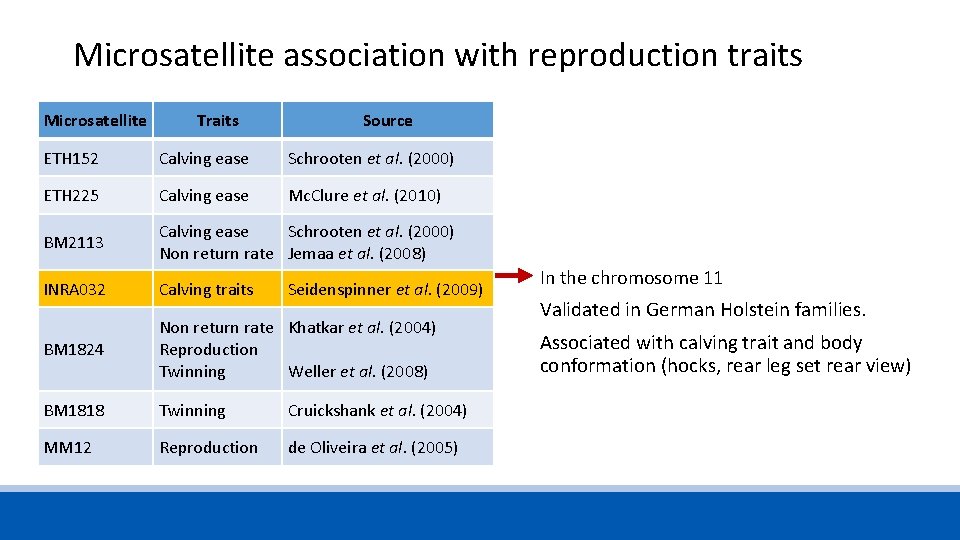
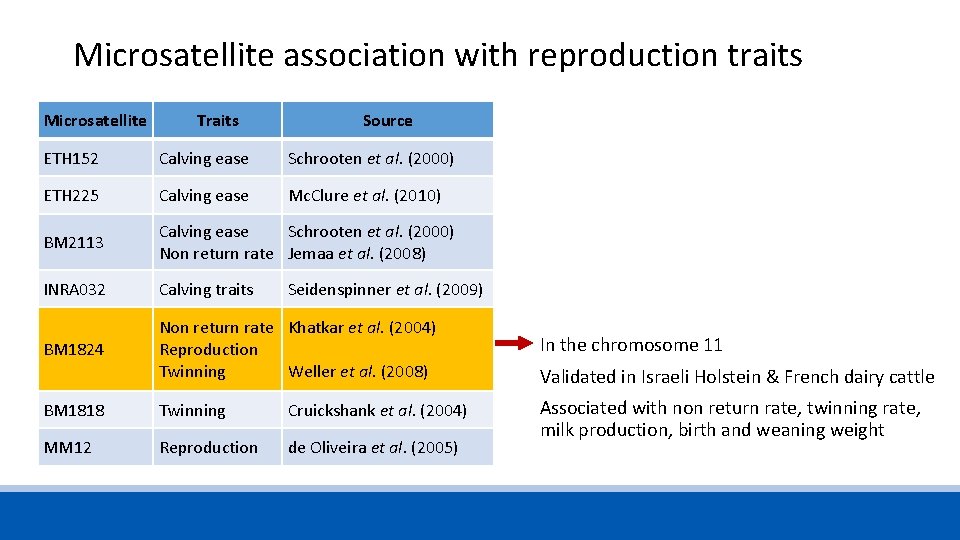
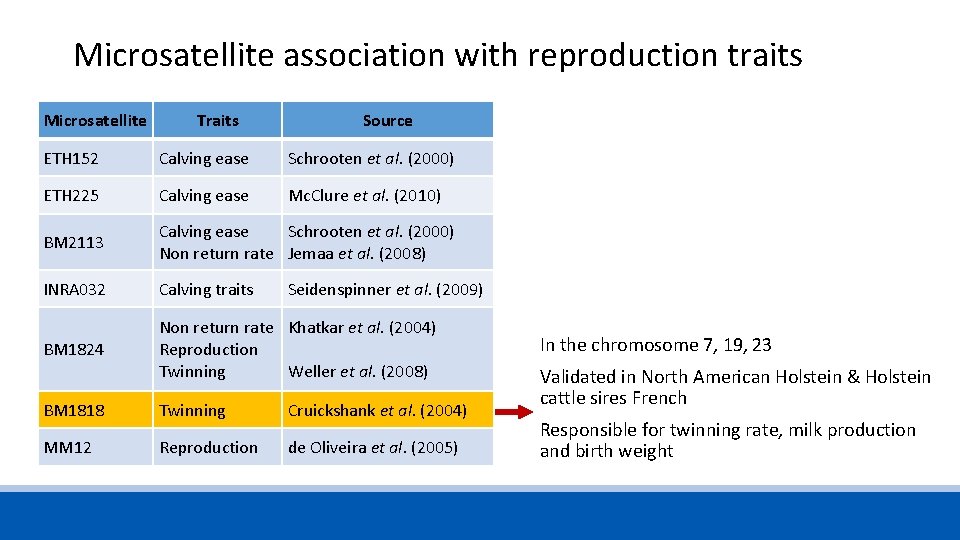
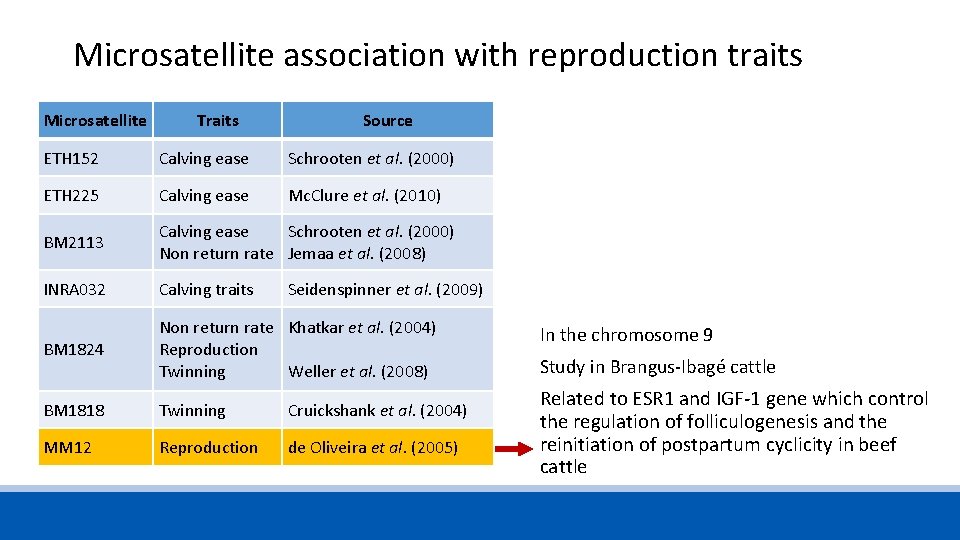
Microsatellite association with reproduction traits Microsatellite Traits Source ETH 152 Calving ease Schrooten et al. (2000) ETH 225 Calving ease Mc. Clure et al. (2010) BM 2113 Calving ease Non return rate Schrooten et al. (2000) Jemaa et al. (2008) INRA 032 Calving traits Seidenspinner et al. (2009) BM 1824 Non return rate Reproduction Twinning Khatkar et al. (2004) BM 1818 Twinning Cruickshank et al. (2004) MM 12 Reproduction de Oliveira et al. (2005) Weller et al. (2008)

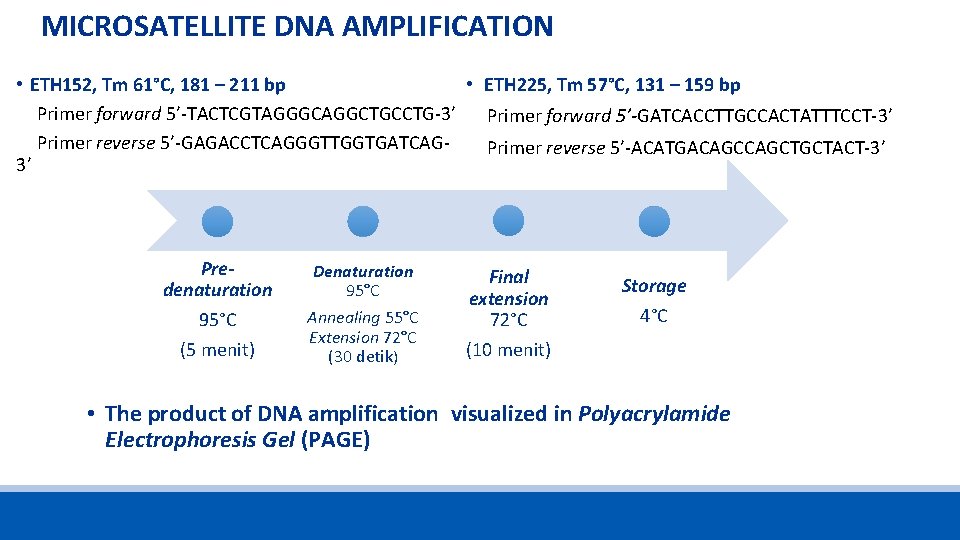
MICROSATELLITE DNA AMPLIFICATION • ETH 225, Tm 57°C, 131 – 159 bp • ETH 152, Tm 61°C, 181 – 211 bp Primer forward 5’-TACTCGTAGGGCAGGCTGCCTG-3’ Primer forward 5’-GATCACCTTGCCACTATTTCCT-3’ Primer reverse 5’-GAGACCTCAGGGTTGGTGATCAGPrimer reverse 5’-ACATGACAGCTGCTACT-3’ 3’ Predenaturation 95°C (5 menit) Denaturation 95°C Annealing 55°C Extension 72°C (30 detik) Final extension 72°C (10 menit) Storage 4°C • The product of DNA amplification visualized in Polyacrylamide Electrophoresis Gel (PAGE)

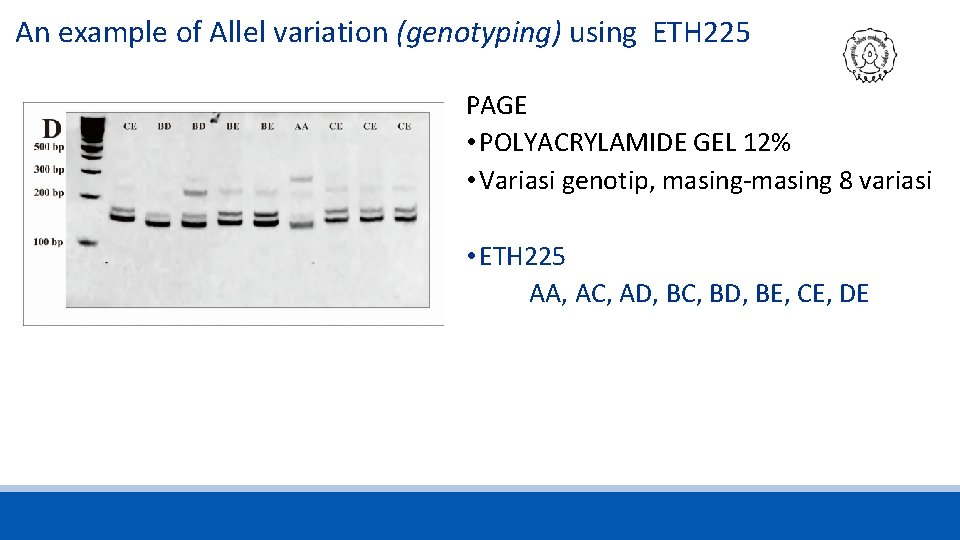
An example of Allel variation (genotyping) using ETH 225 PAGE • POLYACRYLAMIDE GEL 12% • Variasi genotip, masing-masing 8 variasi • ETH 225 AA, AC, AD, BC, BD, BE, CE, DE

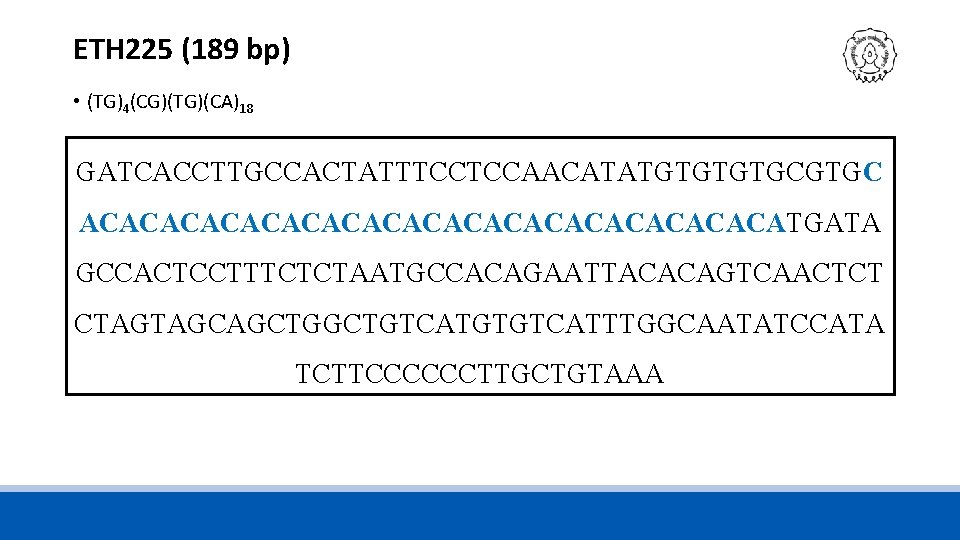
ETH 225 (189 bp) • (TG)4(CG)(TG)(CA)18 GATCACCTTGCCACTATTTCCTCCAACATATGTGCGTGC ACACACACACACACACACATGATA GCCACTCCTTTCTCTAATGCCACAGAATTACACAGTCAACTCT CTAGTAGCAGCTGTCATGTGTCATTTGGCAATATCCATA TCTTCCCCCCTTGCTGTAAA

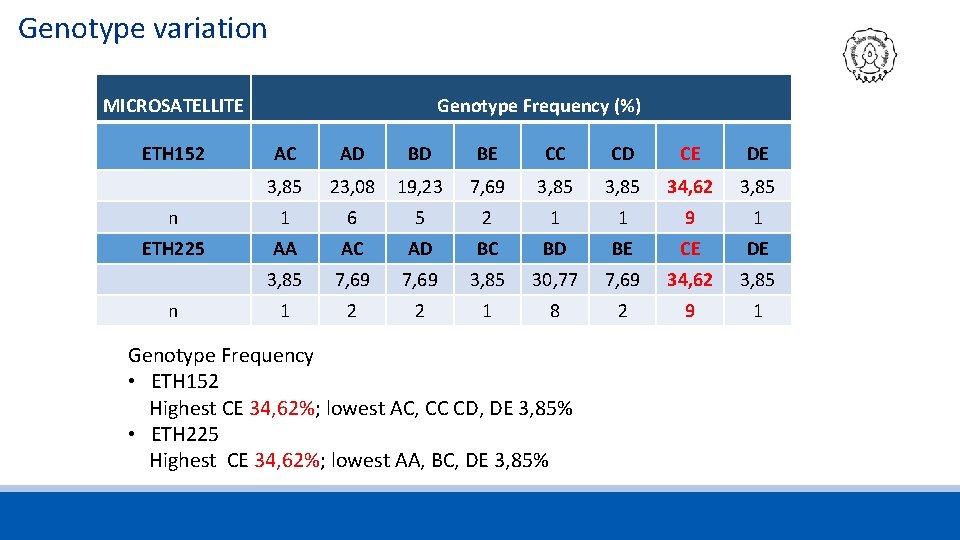
Genotype variation MICROSATELLITE ETH 152 Genotype Frequency (%) AC AD BD BE CC CD CE DE 3, 85 23, 08 19, 23 7, 69 3, 85 34, 62 3, 85 n 1 6 5 2 1 1 9 1 ETH 225 AA AC AD BC BD BE CE DE 3, 85 7, 69 3, 85 30, 77 7, 69 34, 62 3, 85 1 2 2 1 8 2 9 1 n Genotype Frequency • ETH 152 Highest CE 34, 62%; lowest AC, CC CD, DE 3, 85% • ETH 225 Highest CE 34, 62%; lowest AA, BC, DE 3, 85%

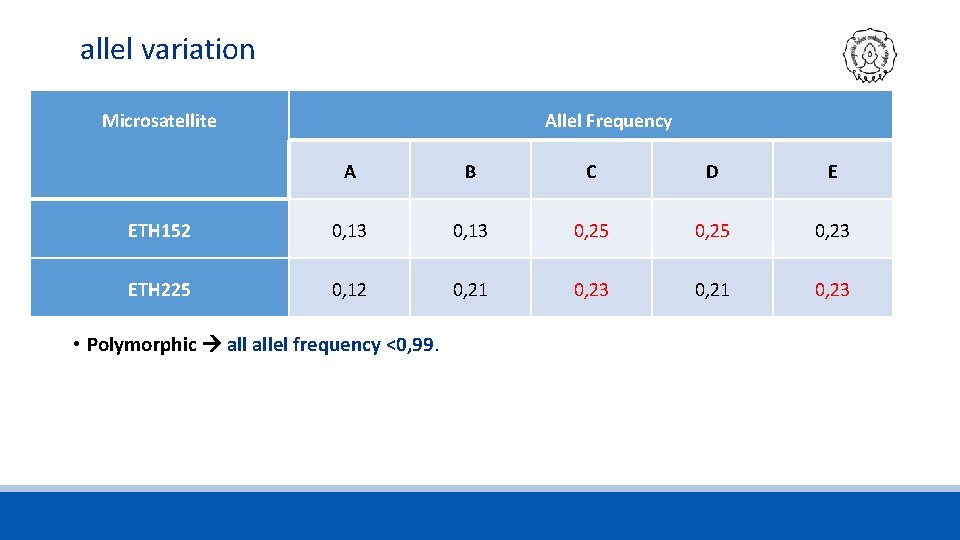
allel variation Microsatellite Allel Frequency A B C D E ETH 152 0, 13 0, 25 0, 23 ETH 225 0, 12 0, 21 0, 23 • Polymorphic allel frequency <0, 99.

• Analyse genotype vs phenotype? • Association?

Other projects: • Sellection of Black rice by irradiation to get a new variety of black rice with shorter period of harvesting time and lowering plant size. • Gene markers for production traits in beef cattle. Work on growth homone gene and mitochondrial DNA • Comparative genetic diversity analysis of local zebu and the Fogera cattle by using microsatellite markers for enhancing milk production and climate change adaptation in selected production and model sites of western Amhara, Ethiopia (Kefyalew Alemayehu (kefyale@gmail. com), Professor Sutarno (nnsutarno@yahoo. com), Dr Sigit Prastowo)

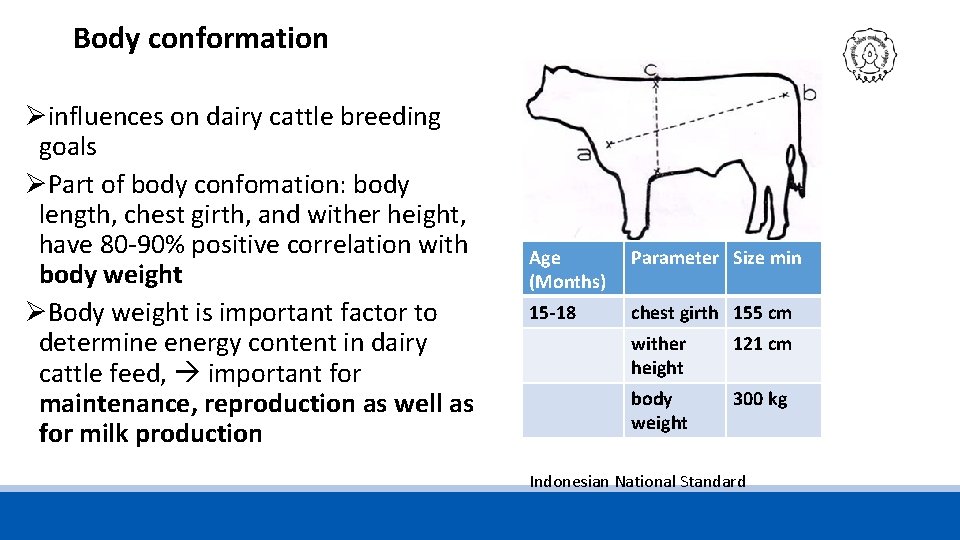
Body conformation Øinfluences on dairy cattle breeding goals ØPart of body confomation: body length, chest girth, and wither height, have 80 -90% positive correlation with body weight ØBody weight is important factor to determine energy content in dairy cattle feed, important for maintenance, reproduction as well as for milk production Age (Months) Parameter Size min 15 -18 chest girth 155 cm wither height 121 cm body weight 300 kg Indonesian National Standard

Microsatellite association with reproduction traits Microsatellite Traits Source ETH 152 Calving ease Schrooten et al. (2000) ETH 225 Calving ease Mc. Clure et al. (2010) BM 2113 Calving ease Schrooten et al. (2000) Non return rate Jemaa et al. (2008) INRA 032 Calving traits BM 1824 Non return rate Khatkar et al. (2004) Reproduction Twinning Weller et al. (2008) BM 1818 Twinning Cruickshank et al. (2004) MM 12 Reproduction de Oliveira et al. (2005) Seidenspinner et al. (2009) In the chromosome 5 Validated in Dutch Holstein-Friesian population Related with calving ease and birth weight

Microsatellite association with reproduction traits Microsatellite Traits Source ETH 152 Calving ease Schrooten et al. (2000) ETH 225 Calving ease Mc. Clure et al. (2010) BM 2113 Calving ease Schrooten et al. (2000) Non return rate Jemaa et al. (2008) INRA 032 Calving traits BM 1824 Non return rate Khatkar et al. (2004) Reproduction Twinning Weller et al. (2008) BM 1818 Twinning Cruickshank et al. (2004) MM 12 Reproduction de Oliveira et al. (2005) Seidenspinner et al. (2009) In the chromosome 9 Validated in commercial Angus cattle Associated with calving ease, birth weight, weaning and yearling weight, milk production

Microsatellite association with reproduction traits Microsatellite Traits Source ETH 152 Calving ease Schrooten et al. (2000) ETH 225 Calving ease Mc. Clure et al. (2010) BM 2113 Calving ease Schrooten et al. (2000) Non return rate Jemaa et al. (2008) In the chromosome 2 INRA 032 Calving traits BM 1824 Non return rate Khatkar et al. (2004) Reproduction Twinning Weller et al. (2008) Suggestive linkage with calving ease, nonreturn post-insemination, milk production and body conformation BM 1818 Twinning Cruickshank et al. (2004) MM 12 Reproduction de Oliveira et al. (2005) Seidenspinner et al. (2009) Validated in Dutch Holstein-Friesian population

Microsatellite association with reproduction traits Microsatellite Traits Source ETH 152 Calving ease Schrooten et al. (2000) ETH 225 Calving ease Mc. Clure et al. (2010) BM 2113 Calving ease Schrooten et al. (2000) Non return rate Jemaa et al. (2008) INRA 032 Calving traits BM 1824 Non return rate Khatkar et al. (2004) Reproduction Twinning Weller et al. (2008) BM 1818 Twinning Cruickshank et al. (2004) MM 12 Reproduction de Oliveira et al. (2005) Seidenspinner et al. (2009) In the chromosome 11 Validated in German Holstein families. Associated with calving trait and body conformation (hocks, rear leg set rear view)

Microsatellite association with reproduction traits Microsatellite Traits Source ETH 152 Calving ease Schrooten et al. (2000) ETH 225 Calving ease Mc. Clure et al. (2010) BM 2113 Calving ease Schrooten et al. (2000) Non return rate Jemaa et al. (2008) INRA 032 Calving traits BM 1824 Non return rate Khatkar et al. (2004) Reproduction Twinning Weller et al. (2008) In the chromosome 11 BM 1818 Twinning Cruickshank et al. (2004) MM 12 Reproduction de Oliveira et al. (2005) Associated with non return rate, twinning rate, milk production, birth and weaning weight Seidenspinner et al. (2009) Validated in Israeli Holstein & French dairy cattle

Microsatellite association with reproduction traits Microsatellite Traits Source ETH 152 Calving ease Schrooten et al. (2000) ETH 225 Calving ease Mc. Clure et al. (2010) BM 2113 Calving ease Schrooten et al. (2000) Non return rate Jemaa et al. (2008) INRA 032 Calving traits BM 1824 Non return rate Khatkar et al. (2004) Reproduction Twinning Weller et al. (2008) Seidenspinner et al. (2009) BM 1818 Twinning Cruickshank et al. (2004) MM 12 Reproduction de Oliveira et al. (2005) In the chromosome 7, 19, 23 Validated in North American Holstein & Holstein cattle sires French Responsible for twinning rate, milk production and birth weight

Microsatellite association with reproduction traits Microsatellite Traits Source ETH 152 Calving ease Schrooten et al. (2000) ETH 225 Calving ease Mc. Clure et al. (2010) BM 2113 Calving ease Schrooten et al. (2000) Non return rate Jemaa et al. (2008) INRA 032 Calving traits BM 1824 Non return rate Khatkar et al. (2004) Reproduction Twinning Weller et al. (2008) BM 1818 Twinning Cruickshank et al. (2004) MM 12 Reproduction de Oliveira et al. (2005) Seidenspinner et al. (2009) In the chromosome 9 Study in Brangus-Ibagé cattle Related to ESR 1 and IGF-1 gene which control the regulation of folliculogenesis and the reinitiation of postpartum cyclicity in beef cattle

Thank You 7/27/19 31