Manual De Novo Peptide MSMS Interpretation For Evaluating



- Slides: 18

Manual De Novo Peptide MS/MS Interpretation For Evaluating Database Search Results Karl R. Clauser Broad Institute of MIT and Harvard Bioinformatics for Protein Identification ASMS Fall Workshop Baltimore, MD November 5 -6, 2009 Karl Clauser Proteomics and Biomarker Discovery 12/22/2021 1

Outline • AA properties • Fragmentation pathways and ion types • b/y pairs • Fragment charge from mass defect • Non-mobile proton • Neutral loss ion types • Mass tolerance units and isobaric AA’s • Other Tutorials Karl Clauser Proteomics and Biomarker Discovery 12/22/2021 2

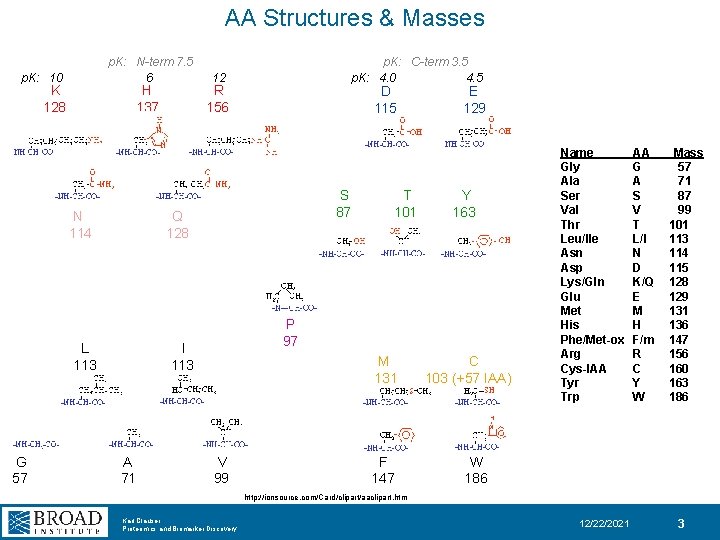
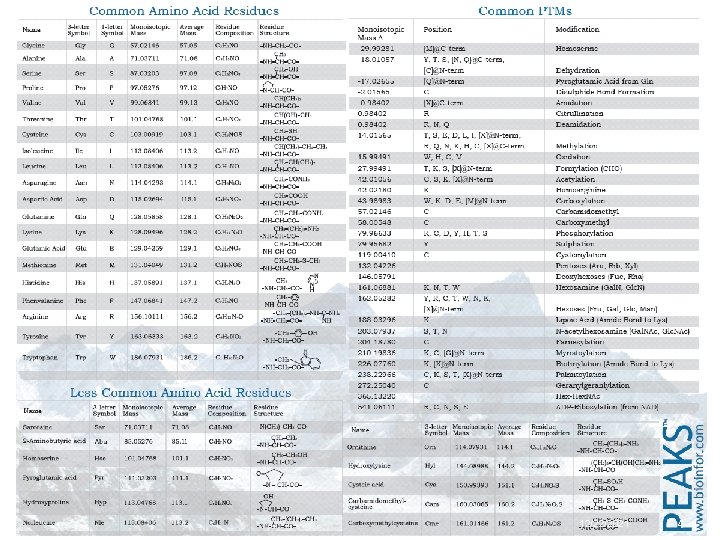
AA Structures & Masses p. K: N-term 7. 5 6 p. K: 10 K 128 H 137 N 114 12 R 156 D 115 S 87 Q 128 L 113 G 57 p. K: C-term 3. 5 p. K: 4. 0 4. 5 Y 163 P 97 I 113 A 71 T 101 E 129 M 131 V 99 F 147 C 103 (+57 IAA) Name Gly Ala Ser Val Thr Leu/Ile Asn Asp Lys/Gln Glu Met His Phe/Met-ox Arg Cys-IAA Tyr Trp AA G A S V T L/I N D K/Q E M H F/m R C Y W Mass 57 71 87 99 101 113 114 115 128 129 131 136 147 156 160 163 186 W 186 http: //ionsource. com/Card/clipart/aaclipart. htm Karl Clauser Proteomics and Biomarker Discovery 12/22/2021 3

12/22/2021 4

12/22/2021 5

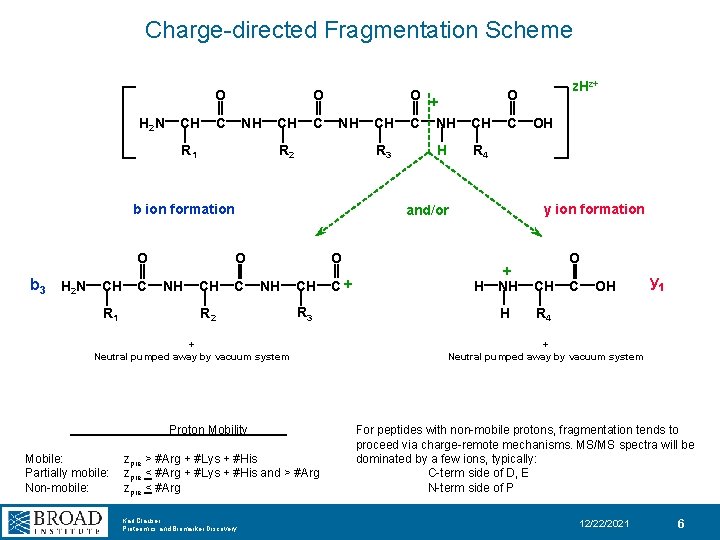
Charge-directed Fragmentation Scheme O H 2 N CH O C NH R 1 CH C NH R 2 R 3 b ion formation O O R 2 R 3 Proton Mobility zpre > #Arg + #Lys + #His zpre < #Arg + #Lys + #His and > #Arg zpre < #Arg Karl Clauser Proteomics and Biomarker Discovery CH C OH R 4 y ion formation O + Neutral pumped away by vacuum system Mobile: Partially mobile: Non-mobile: H z. Hz+ O and/or b 3 H 2 N CH C NH CH C + R 1 CH O + C NH H O + NH CH H R 4 C OH y 1 + Neutral pumped away by vacuum system For peptides with non-mobile protons, fragmentation tends to proceed via charge-remote mechanisms. MS/MS spectra will be dominated by a few ions, typically: C-term side of D, E N-term side of P 12/22/2021 6

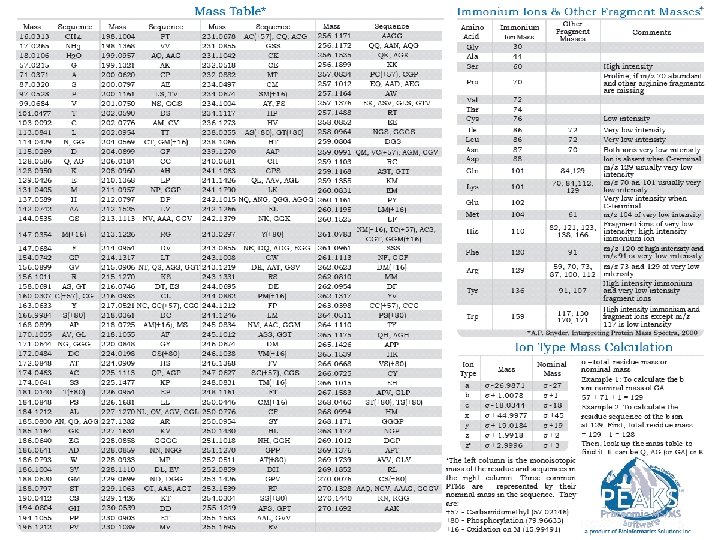
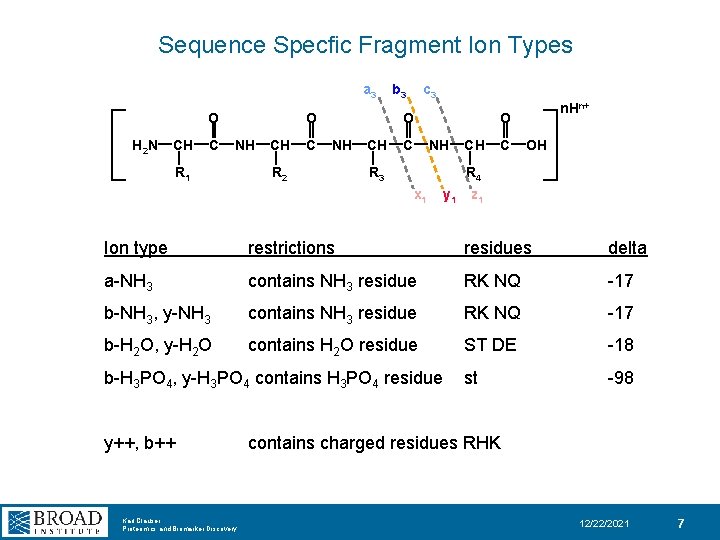
Sequence Specfic Fragment Ion Types a 3 O H 2 N CH C O NH R 1 CH C b 3 c 3 O NH R 2 CH C n. Hn+ O NH R 3 CH C OH R 4 x 1 y 1 z 1 Ion type restrictions residues delta a-NH 3 contains NH 3 residue RK NQ -17 b-NH 3, y-NH 3 contains NH 3 residue RK NQ -17 b-H 2 O, y-H 2 O contains H 2 O residue ST DE -18 st -98 b-H 3 PO 4, y-H 3 PO 4 contains H 3 PO 4 residue y++, b++ Karl Clauser Proteomics and Biomarker Discovery contains charged residues RHK 12/22/2021 7

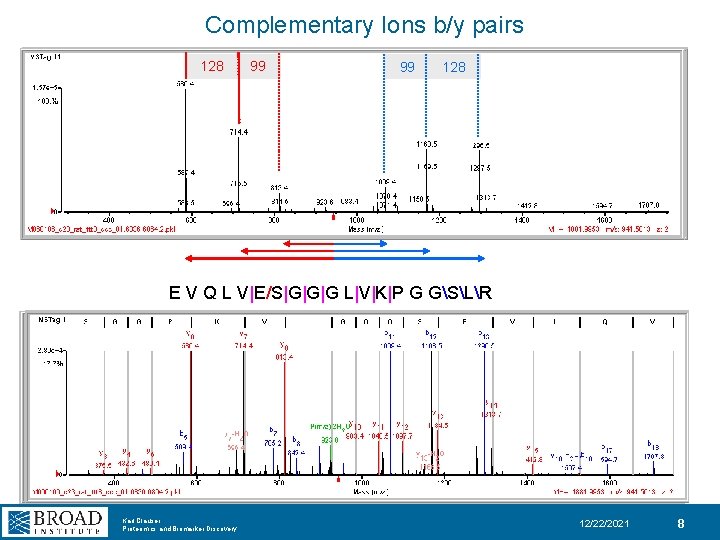
Complementary Ions b/y pairs 128 99 99 128 E V Q L V|E/S|G|G|G L|V|K|P G GSLR Karl Clauser Proteomics and Biomarker Discovery 12/22/2021 8

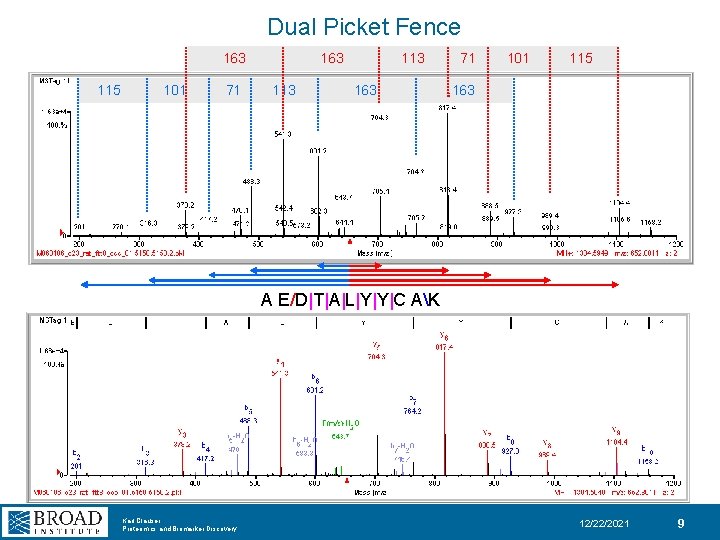
Dual Picket Fence 163 115 101 71 163 113 163 71 101 115 163 A E/D|T|A|L|Y|Y|C AK Karl Clauser Proteomics and Biomarker Discovery 12/22/2021 9

Uniqueness of a Peptide Sequence Clauser, K. R. ; Baker, P. R. ; Burlingame, A. L. " Role of Accurate Mass Measurement ( +/- 10 ppm) in Protein Identification Strategies Employing MS or MS/MS and Database Searching", Anal. Chem. 1999, 71, 2871 -2882. Karl Clauser Proteomics and Biomarker Discovery 12/22/2021 10

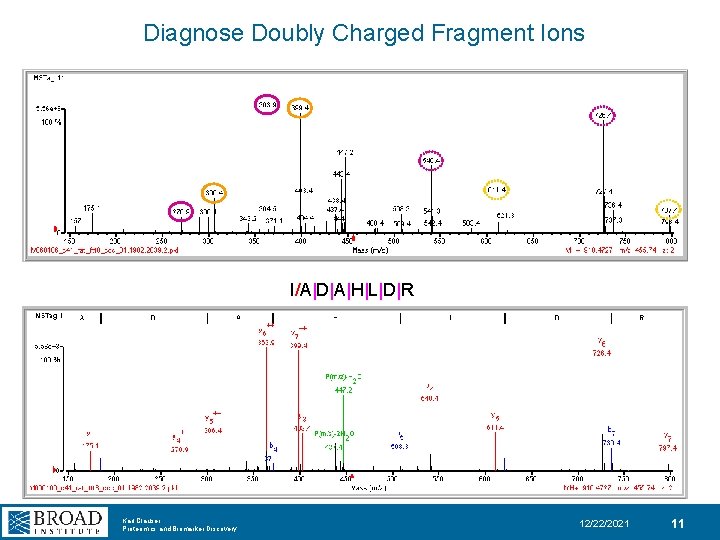
Diagnose Doubly Charged Fragment Ions I/A|D|A|H|L|D|R Karl Clauser Proteomics and Biomarker Discovery 12/22/2021 11

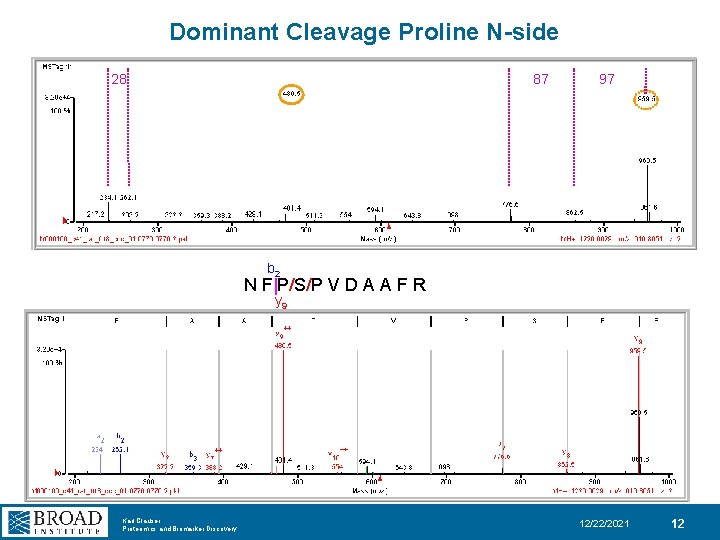
Dominant Cleavage Proline N-side 28 87 97 b 2 N F|P/S/P V D A A F R y 9 Karl Clauser Proteomics and Biomarker Discovery 12/22/2021 12

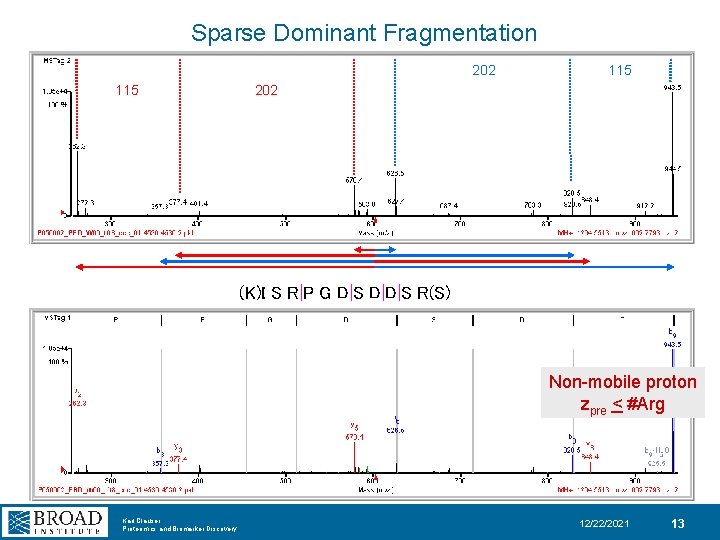
Sparse Dominant Fragmentation 202 115 202 (K)I S R|P G D|S D|D|S R(S) Non-mobile proton zpre < #Arg Karl Clauser Proteomics and Biomarker Discovery 12/22/2021 13

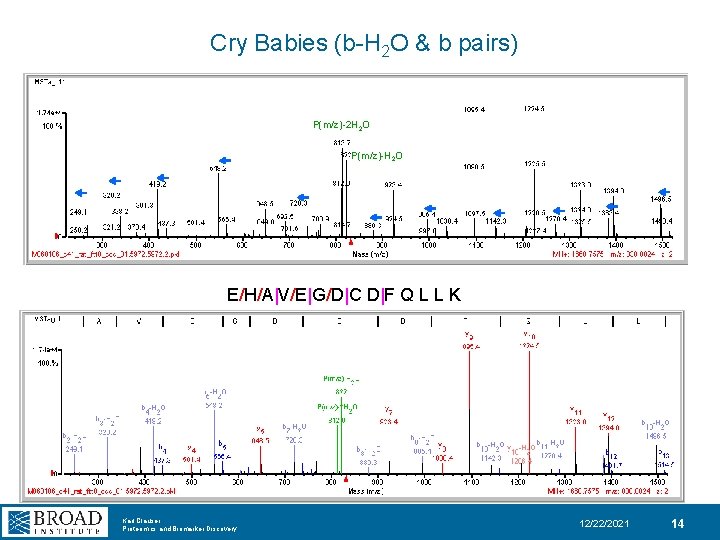
Cry Babies (b-H 2 O & b pairs) P(m/z)-2 H 2 O P(m/z)-H 2 O E/H/A|V/E|G/D|C D|F Q L L K Karl Clauser Proteomics and Biomarker Discovery 12/22/2021 14

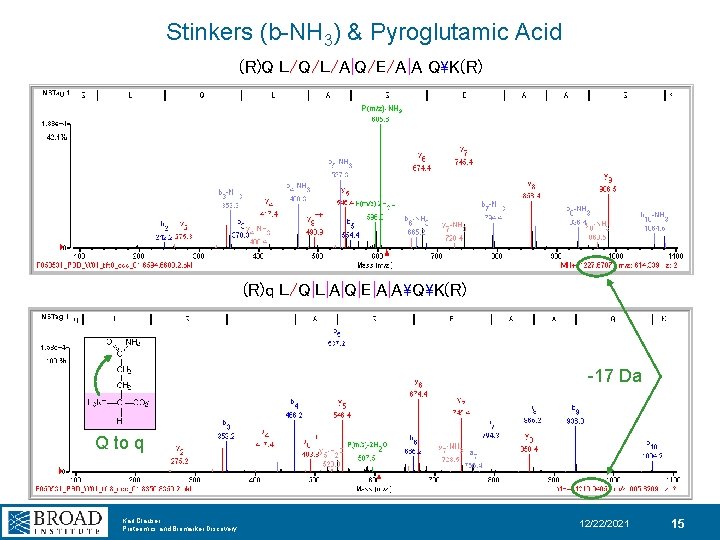
Stinkers (b-NH 3) & Pyroglutamic Acid (R)Q L/Q/L/A|Q/E/A|A QK(R) P(m/z)-NH 3 (R)q L/Q|L|A|Q|E|A|AQK(R) -17 Da Q to q Karl Clauser Proteomics and Biomarker Discovery 12/22/2021 15

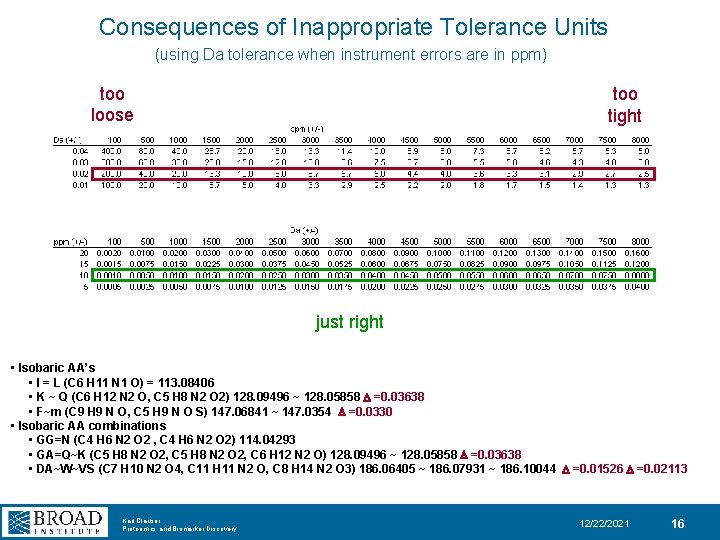
Consequences of Inappropriate Tolerance Units (using Da tolerance when instrument errors are in ppm) too loose too tight just right • Isobaric AA’s • I = L (C 6 H 11 N 1 O) = 113. 08406 • K ~ Q (C 6 H 12 N 2 O, C 5 H 8 N 2 O 2) 128. 09496 ~ 128. 05858 D =0. 03638 • F~m (C 9 H 9 N O, C 5 H 9 N O S) 147. 06841 ~ 147. 0354 D =0. 0330 • Isobaric AA combinations • GG=N (C 4 H 6 N 2 O 2 , C 4 H 6 N 2 O 2) 114. 04293 • GA=Q~K (C 5 H 8 N 2 O 2, C 6 H 12 N 2 O) 128. 09496 ~ 128. 05858 D =0. 03638 • DA~W~VS (C 7 H 10 N 2 O 4, C 11 H 11 N 2 O, C 8 H 14 N 2 O 3) 186. 06405 ~ 186. 07931 ~ 186. 10044 D =0. 01526 D =0. 02113 Karl Clauser Proteomics and Biomarker Discovery 12/22/2021 16

Additional Resources Google: “de novo sequencing tutorial” Don Hunt and Jeff Shabanowitz - manual http: //www. ionsource. com/tutorial/De. Novo. TOC. htm Rich Johnson - manual http: //www. abrf. org/Research. Groups/Mass. Spectrometry/EPosters/ms 97 quiz/Sequencing. Tutorial. html PEAKS - automated http: //www. bioinformaticssolutions. com/products/peaks/support/tutorials/PEAKS_De_Novo. html Karl Clauser Proteomics and Biomarker Discovery 12/22/2021 17

Acknowledgements Broad Institute MIT Steve Carr Terri Addona Jinyan Du Michael Yaffe Majbrit Hjerrld Drew Lowery Karl Clauser Proteomics and Biomarker Discovery 12/22/2021 18