Linear Mixed Models for Longitudinal and Clustered Data








- Slides: 16

Linear Mixed Models for Longitudinal and Clustered Data 4/20/2021 Daniel Zhao, Ph. D Department of Biostatistics and Epidemiology Hudson College of Public Health University of Oklahoma Health Sciences Center

Outline • Lecture (45 min) • • • Data Model set up Estimation Prediction Interpretation • R demo (30 min) • lme 4 R package • In-class exercise (45 min) 2

Typical data suitable for LMM 1. Clustered Data (can be cross-sectional) 1. A cardiovascular study conducted on a group of families (clusters). 2. Outcomes (blood pressures) of patients from different families are independent, but from the same families are correlated. 2. Longitudinal Data (sequential in time) 1. A clinical trial to compare two medications on treating hypertension 2. Outcomes (blood pressures) from different patients are independent, but from the same patients are correlated. Note: Classical linear regression models ignoring the correlation structure will produce biased parameter estimates. 3

Example: Treatment of Lead-Exposed Children Trial • Exposure to lead during infancy is associated with substantial deficits in tests of cognitive ability • Chelation treatment of children with high lead levels usually requires injections and hospitalization • A new agent, Succimer, can be given orally • Randomized trial examining changes in blood lead level during course of treatment • 100 children randomized to placebo or Succimer • Measures of blood lead level at baseline, 1, 4 and 6 weeks 4

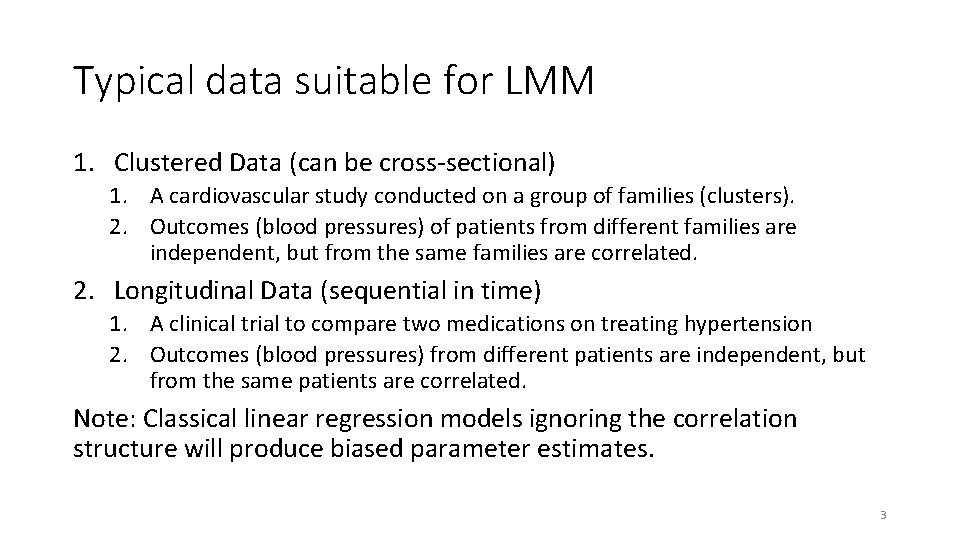
Table 1: Blood lead levels (μg/d. L) at baseline, week 1, week 4, and week 6 for 8 randomly selected children. 5

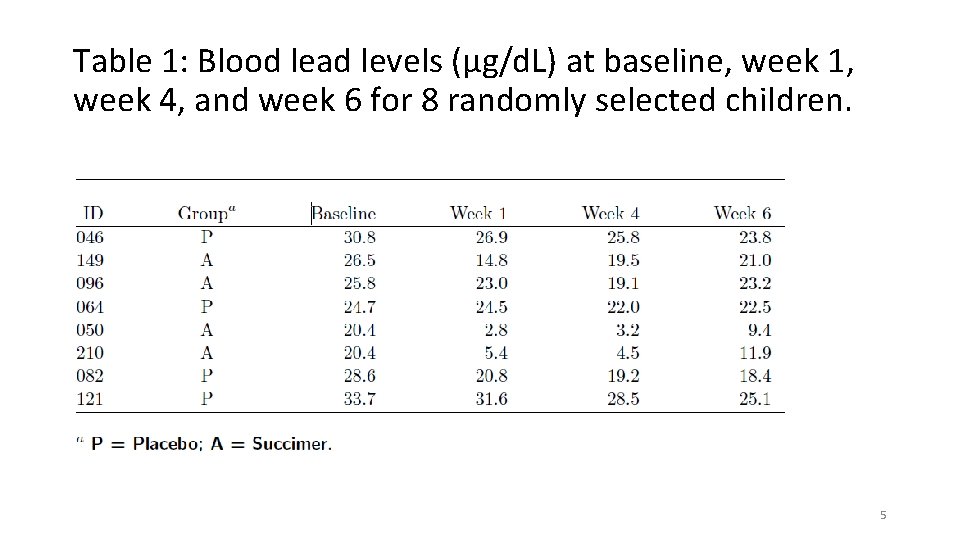
Table 2: Mean blood lead levels (and standard deviation) at baseline, week 1, week 4, and week 6. 6

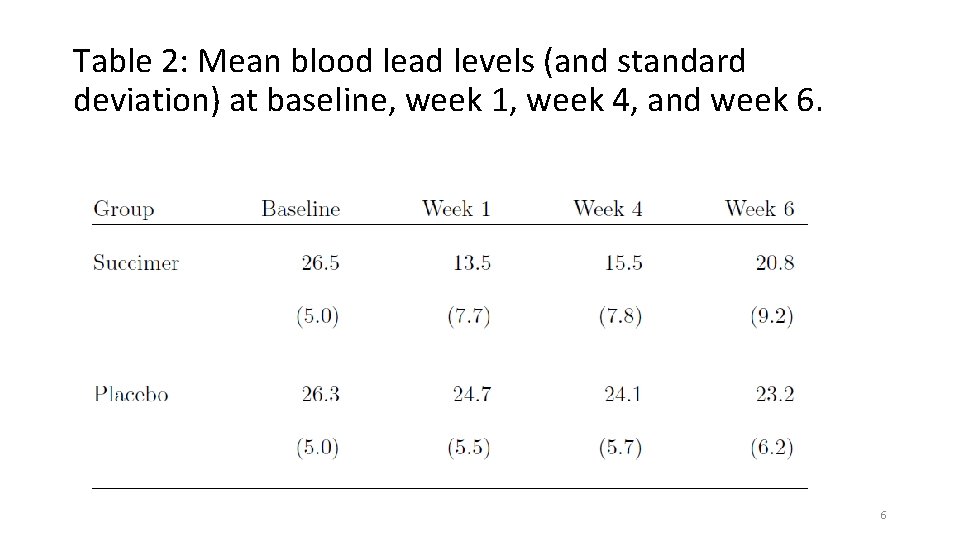
Figure 1: Spaghetti plot of blood lead levels at baseline, week 1, week 4, and week 6 7

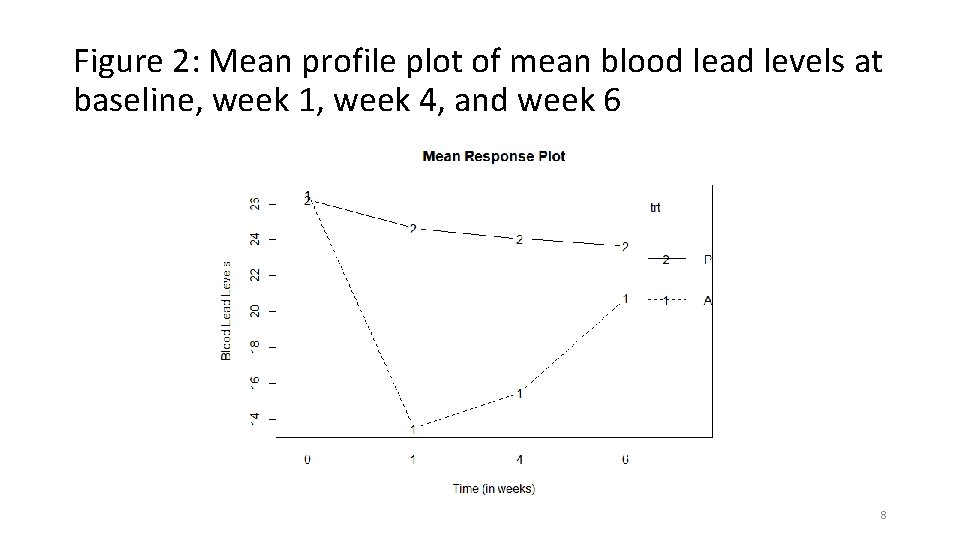
Figure 2: Mean profile plot of mean blood lead levels at baseline, week 1, week 4, and week 6 8

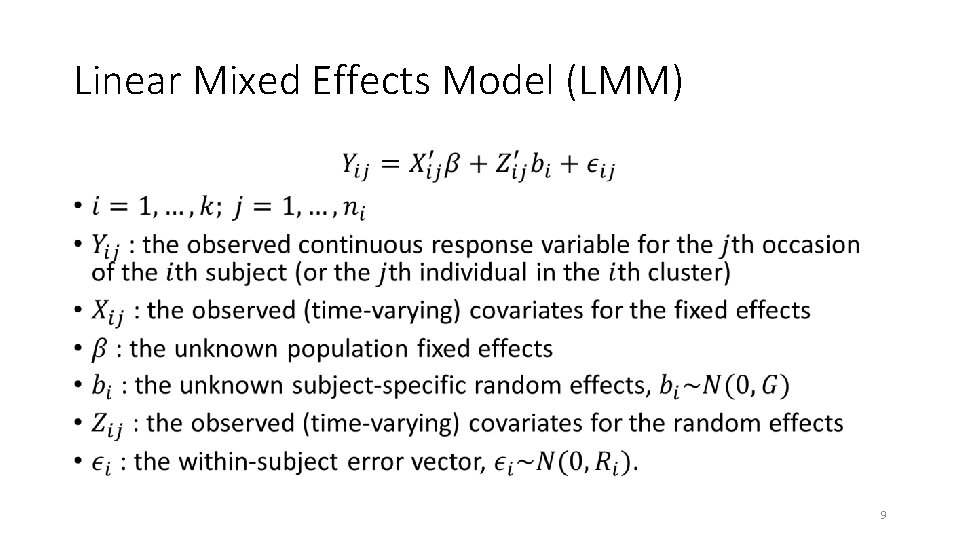
Linear Mixed Effects Model (LMM) • 9

Mean and Variance • 10

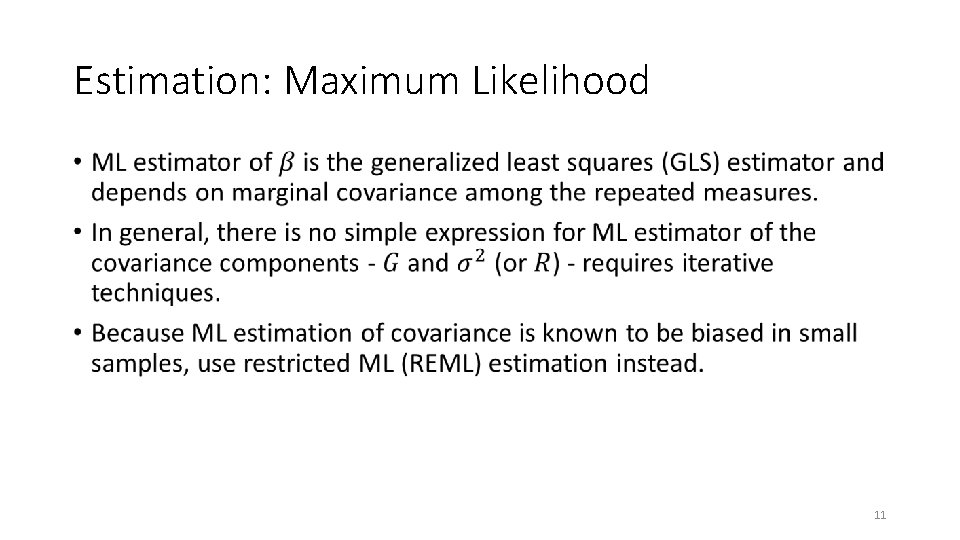
Estimation: Maximum Likelihood • 11

Prediction • 12

Model Selection for LMM • Both the mean model and covariance structure need to be selected • A practical approach is to fit a saturated mean model and use AIC or BIC to pick the best covariance structure. • Using the chosen covariance structure, find the final mean model through backward elimination 13

Handling of time in longitudinal studies • If the time points for everyone are the same (say, weeks 1, 3, 4, 5, etc) • Can treat time as a continuous variable if response is linear in time or functional form can be modeled • Otherwise, treat time as a categorical variable and specify a reference time • If the time points for everyone are not the same • Time can only be treated as continuous variable 14

R packages for LMM • foreign package, gls function: less often used. • nlme package, nlme function: can be used for nonlinear mixed effects models • lme 4 package, lmer function: most commonly used 15

Syntax of lmer function in lme 4 package • In lmer the model is specified by the formula argument. As in most R model-fitting functions, this is the first argument. • The model formula consists of two expressions separated by the ~ symbol. • The expression on the left of ~ is the response variable. • The RHS consists of one or more terms separated by `+' symbols. • A random-effects term consists of two expressions separated by the vertical bar |. • The expression on the right of the `|' is evaluated as a factor, which we call the grouping factor for that term. 16