Integrating Phenotypic Data With Genomic Genetic and Genotypic






- Slides: 13

Integrating Phenotypic Data With Genomic, Genetic and Genotypic Data Using Chado Sook Jung, Taein Lee, Stephen Ficklin, Jing Yu, Dorrie Main

Outline § Introduction of GDR and Cotton. Gen § Chado the generic schema § Storing Stock Data § Storing Phenotypic Data (trait, dataset, etc) § Storing Genotypic Data § Integration with genetic and genomic Data § Conclusion

Database projects of Main lab § Major databases with genomic, genetic, phenotypic and genotypic data 1. GDR: Genome Database for Rosaceae Genomic. Gemetoc and Breeding data (Private data and data from Ros. Breed project) 2. § • Fruit and Nut, Sat, 12 PM • Computer Demo, Mon, 1: 35 PM • P 0946, Ros. Breed BIM System, Mon, 10 -11: 30 AM Cotton. Gen: Replaced Cotton. DB and Cotton Marker Database • Cotton Genome Initiative, Sun, 3: 50 PM • Computer Demo, Mon, 1: 50 PM Other databases: § Citrus Genome Database, Cool season food legume database, Genome database for Vacciniium § Built using Chado schema and Tripal (Drupal front end for Chado) § Tripal presentation, GMOD workshop, Wed 11: 50 AM

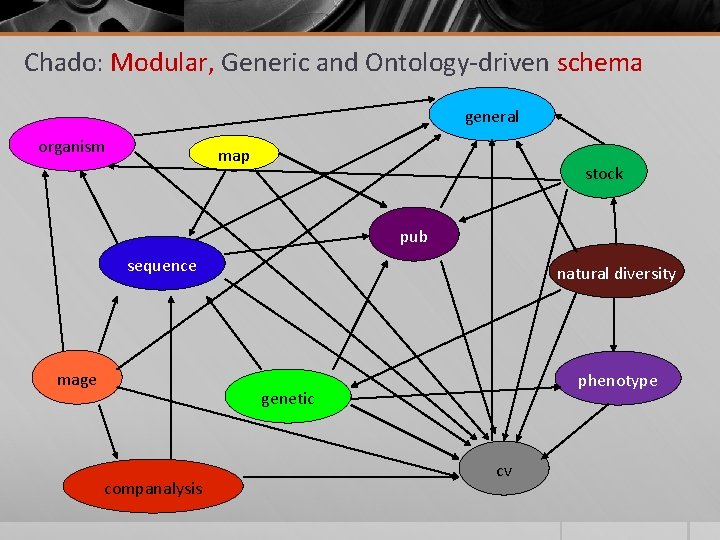
Chado: Modular, Generic and Ontology-driven schema general organism map stock pub sequence mage natural diversity phenotype genetic companalysis cv

Publication

Chado: Modular, Generic and Ontology-driven schema Feature_relationship_id Subject_id Object_id Type_id Abc-m. RNA part_of Abc-gene id ct_ e j ub S o _id ct bje Feature_id Name Uniquename Type_id Organism_id residues gene, m. RNA, marker, QTL, etc Featureprop_id Feature_id Type_id Value rank Repeat_motif Product_size cvterm_id Name definition cv_id Dbxref_id cv cv_id Name definition Sequence Ontology, Gene Ontology, etc

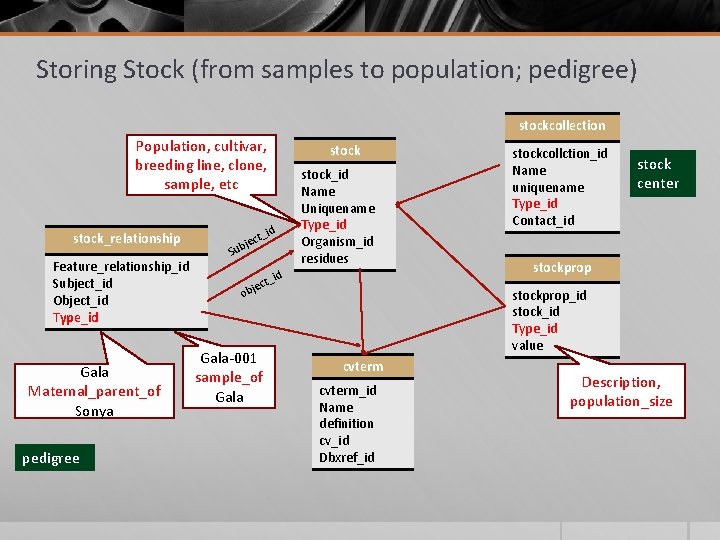
Storing Stock (from samples to population; pedigree) stockcollection Population, cultivar, breeding line, clone, sample, etc stock_relationship Feature_relationship_id Subject_id Object_id Type_id Gala Maternal_parent_of Sonya pedigree ect ubj _id S stock_id Name Uniquename Type_id Organism_id residues t_id ec obj Gala-001 sample_of Gala stockcollction_id Name uniquename Type_id Contact_id stock center stockprop_id stock_id Type_id value cvterm_id Name definition cv_id Dbxref_id Description, population_size

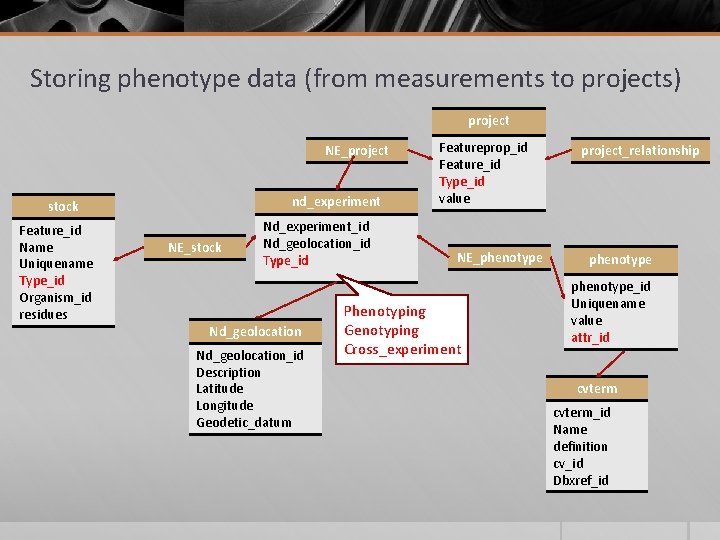
Storing phenotype data (from measurements to projects) project NE_project nd_experiment stock Feature_id Name Uniquename Type_id Organism_id residues NE_stock Nd_experiment_id Nd_geolocation_id Type_id Nd_geolocation_id Description Latitude Longitude Geodetic_datum Featureprop_id Feature_id Type_id value NE_phenotype Phenotyping Genotyping Cross_experiment project_relationship phenotype_id Uniquename value attr_id cvterm_id Name definition cv_id Dbxref_id

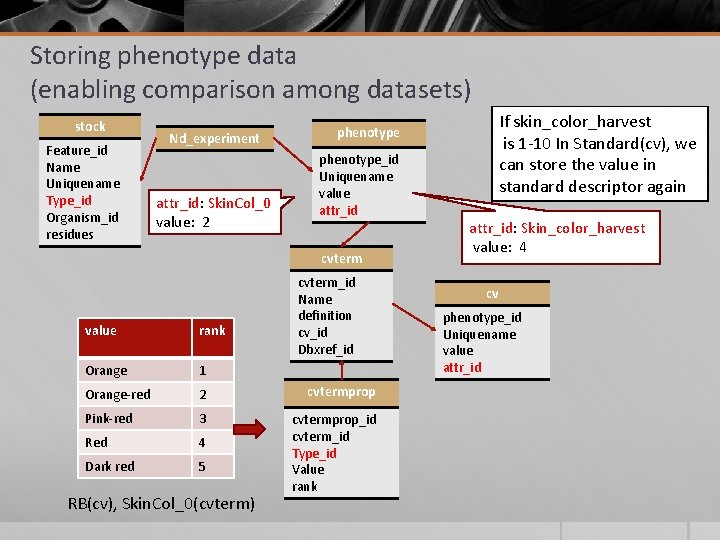
Storing phenotype data (enabling comparison among datasets) stock Feature_id Name Uniquename Type_id Organism_id residues Nd_experiment attr_id: Skin. Col_0 value: 2 phenotype_id Uniquename value attr_id cvterm_id Name definition cv_id Dbxref_id value rank Orange 1 Orange-red 2 cvtermprop Pink-red 3 Red 4 Dark red 5 cvtermprop_id cvterm_id Type_id Value rank RB(cv), Skin. Col_0(cvterm) If skin_color_harvest is 1 -10 In Standard(cv), we can store the value in standard descriptor again phenotype attr_id: Skin_color_harvest value: 4 cv phenotype_id Uniquename value attr_id

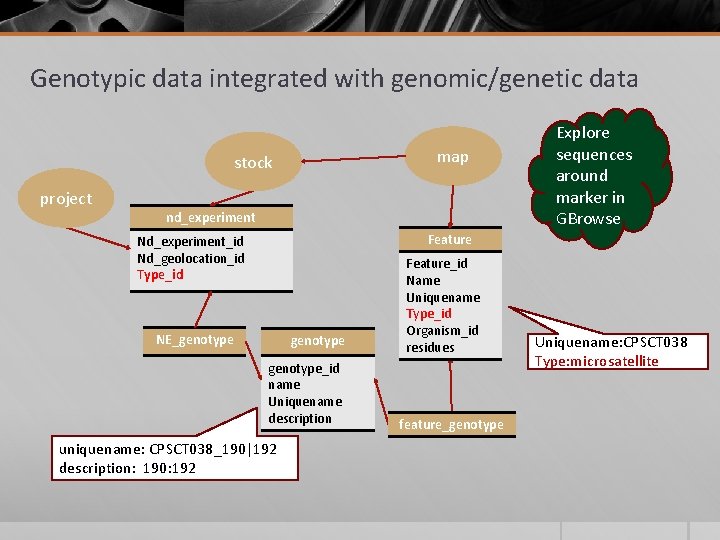
Genotypic data integrated with genomic/genetic data map stock project nd_experiment Explore sequences around marker in GBrowse Feature Nd_experiment_id Nd_geolocation_id Type_id NE_genotype_id name Uniquename description uniquename: CPSCT 038_190|192 description: 190: 192 Feature_id Name Uniquename Type_id Organism_id residues feature_genotype Uniquename: CPSCT 038 Type: microsatellite

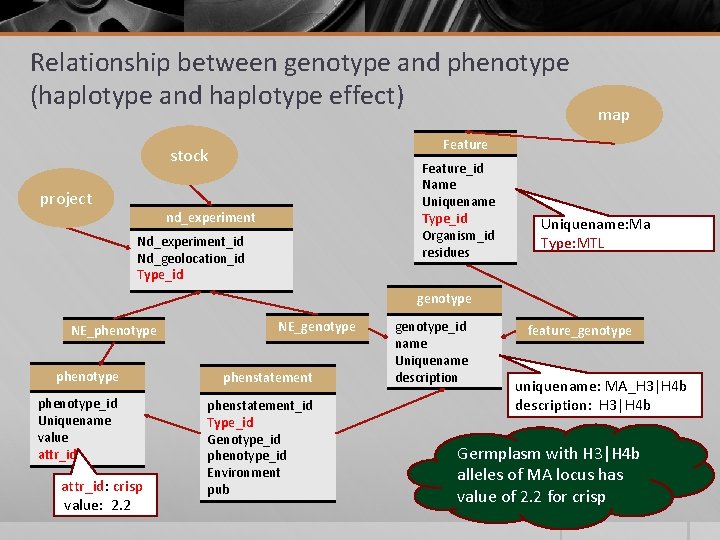
Relationship between genotype and phenotype (haplotype and haplotype effect) Feature stock project map Feature_id Name Uniquename Type_id Organism_id residues nd_experiment Nd_experiment_id Nd_geolocation_id Type_id Uniquename: Ma Type: MTL genotype NE_phenotype NE_genotype phenstatement phenotype_id Uniquename value attr_id phenstatement_id Type_id Genotype_id phenotype_id Environment pub attr_id: crisp value: 2. 2 genotype_id name Uniquename description feature_genotype uniquename: MA_H 3|H 4 b description: H 3|H 4 b Germplasm with H 3|H 4 b alleles of MA locus has value of 2. 2 for crisp

Conclusion § Flexibility and generic characteristic of Chado enables us to store and integrate complex biological data from widely different projects and species § The ontology-driven characteristic makes adding new data types relatively easy. § Performance issue mostly resolved by the use of materialized views

Acknowledgement § Natural diversity module working group Naama Menda, Seth Redmond, Robert M. Buels, Maren Friesen, Yuri Bendana, Lacey. Anne Sanderson, Hilmar Lapp, Taein Lee, Bob Mac. Callum, Kirstin E. Bett, Scott Cain, Dave Clements, Lukas A. Mueller and Dorrie Main § Main Lab team Dorrie Main Taein Lee Stephen Ficklin Jing Yu Chun-Huai Cheng Ping Zheng Anna Blenda Sushan Ru § All Project Co. PIs (tf. GDR, Ros. Breed and Cotton. Gen) § Funding Sources USDA NIFA SCRI, NSF Plant Genome Program, USDA-ARS, Washington Tree Fruit Research Commission, Cotton Incorporated, Washington State University, Clemson University, University of Florida, Boyce Thompson Institute, North Carolina State University