How do we find a knockout for AT



- Slides: 11

How do we find a knockout for AT 4 G 37790 and what is this gene’s function in seed development? A Research Talk by Ita Nagy HC 70 AL Spring 2004

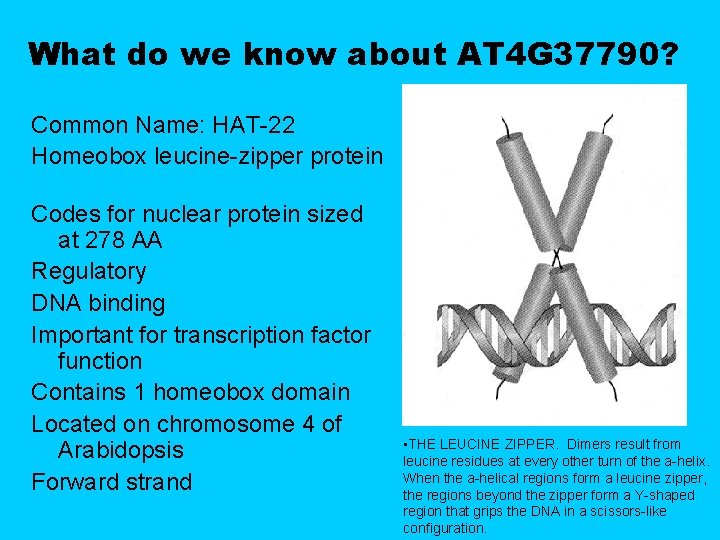
What do we know about AT 4 G 37790? Common Name: HAT-22 Homeobox leucine-zipper protein Codes for nuclear protein sized at 278 AA Regulatory DNA binding Important for transcription factor function Contains 1 homeobox domain Located on chromosome 4 of Arabidopsis Forward strand • THE LEUCINE ZIPPER. Dimers result from leucine residues at every other turn of the a-helix. When the a-helical regions form a leucine zipper, the regions beyond the zipper form a Y-shaped region that grips the DNA in a scissors-like configuration.

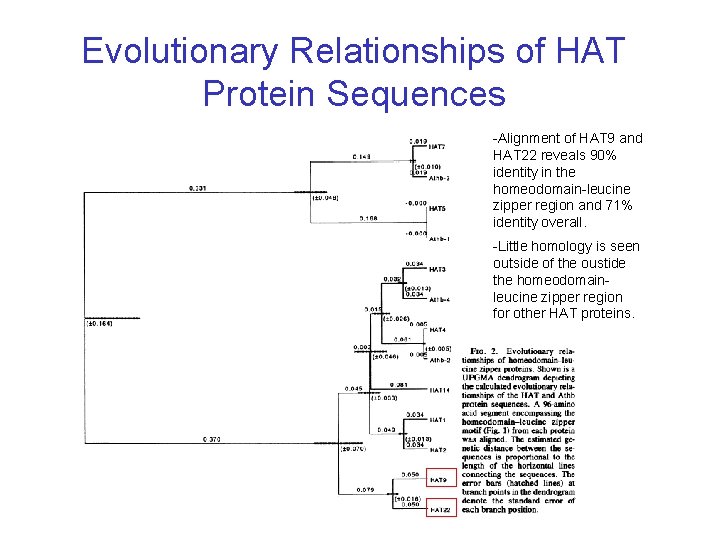
Evolutionary Relationships of HAT Protein Sequences -Alignment of HAT 9 and HAT 22 reveals 90% identity in the homeodomain-leucine zipper region and 71% identity overall. -Little homology is seen outside of the oustide the homeodomainleucine zipper region for other HAT proteins.

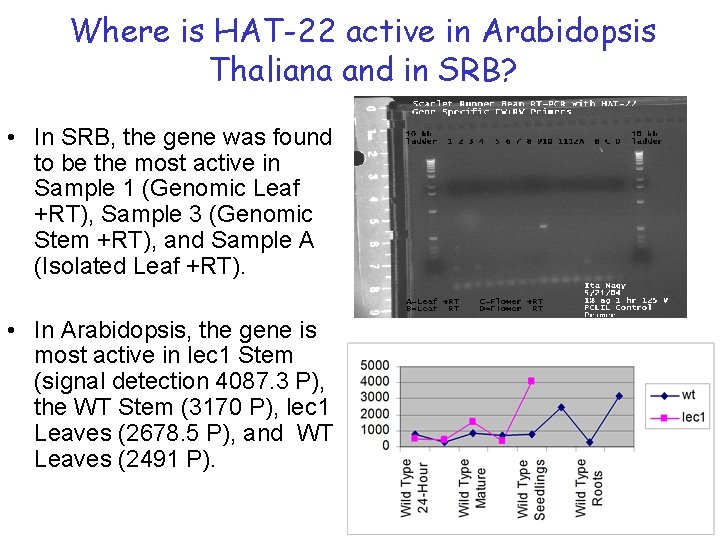
Where is HAT-22 active in Arabidopsis Thaliana and in SRB? • In SRB, the gene was found to be the most active in Sample 1 (Genomic Leaf +RT), Sample 3 (Genomic Stem +RT), and Sample A (Isolated Leaf +RT). • In Arabidopsis, the gene is most active in lec 1 Stem (signal detection 4087. 3 P), the WT Stem (3170 P), lec 1 Leaves (2678. 5 P), and WT Leaves (2491 P).

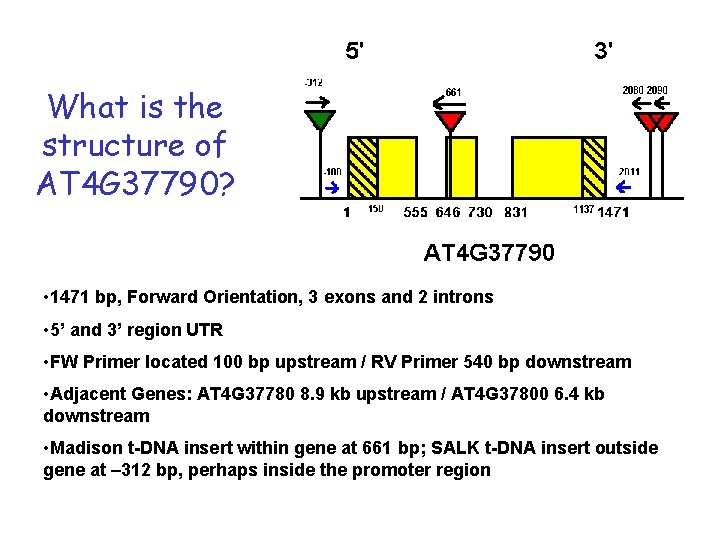
What is the structure of AT 4 G 37790? • 1471 bp, Forward Orientation, 3 exons and 2 introns • 5’ and 3’ region UTR • FW Primer located 100 bp upstream / RV Primer 540 bp downstream • Adjacent Genes: AT 4 G 37780 8. 9 kb upstream / AT 4 G 37800 6. 4 kb downstream • Madison t-DNA insert within gene at 661 bp; SALK t-DNA insert outside gene at – 312 bp, perhaps inside the promoter region

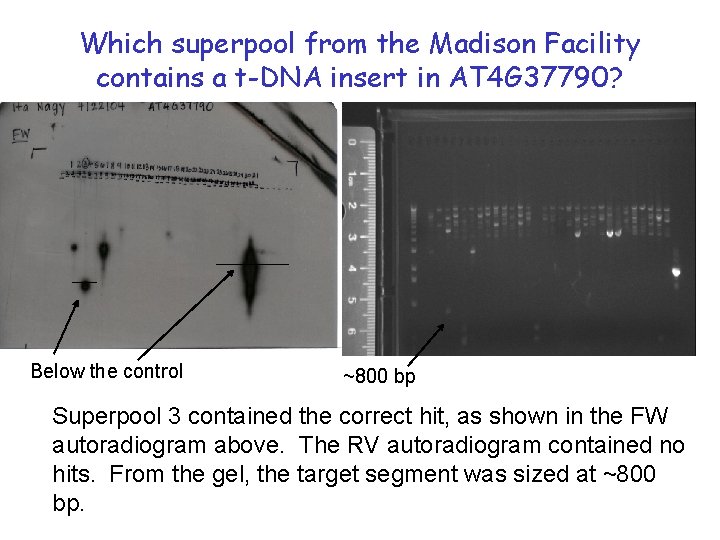
Which superpool from the Madison Facility contains a t-DNA insert in AT 4 G 37790? Below the control ~800 bp Superpool 3 contained the correct hit, as shown in the FW autoradiogram above. The RV autoradiogram contained no hits. From the gel, the target segment was sized at ~800 bp.

Which DNA pool contains a t. DNA insert in AT 4 G 37790? 800 bp • Amplify PCR products from 9 DNA pools belonging to Superpool 3. • Use FW & JL 202 primers and a + control, Superpool 3. • Can we find a PCR product with the same size as the hit segment from our superpool screening (~800 bp)? • The primers amplify an ~800 bp segment in DNA Pool 4.

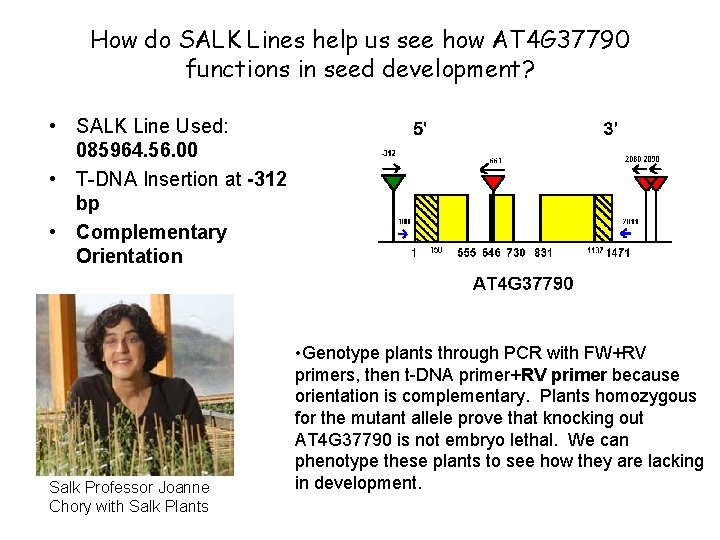
How do SALK Lines help us see how AT 4 G 37790 functions in seed development? • SALK Line Used: 085964. 56. 00 • T-DNA Insertion at -312 bp • Complementary Orientation Salk Professor Joanne Chory with Salk Plants • Genotype plants through PCR with FW+RV primers, then t-DNA primer+RV primer because orientation is complementary. Plants homozygous for the mutant allele prove that knocking out AT 4 G 37790 is not embryo lethal. We can phenotype these plants to see how they are lacking in development.

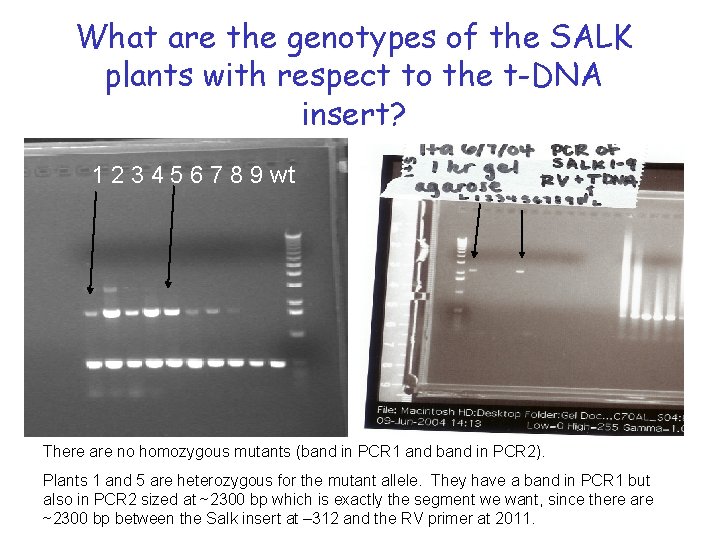
What are the genotypes of the SALK plants with respect to the t-DNA insert? 1 2 3 4 5 6 7 8 9 wt There are no homozygous mutants (band in PCR 1 and band in PCR 2). Plants 1 and 5 are heterozygous for the mutant allele. They have a band in PCR 1 but also in PCR 2 sized at ~2300 bp which is exactly the segment we want, since there are ~2300 bp between the Salk insert at – 312 and the RV primer at 2011.

Where do we go from here? • SALK LINES: Collect seeds from heterozygous mutant plants and attempt to raise homozygous mutants. • If homozygous mutants are obtained, knockout is not embryo lethal. However, we can PHENOTYPE the plants to see whether abnormalities give us clues to the importance of AT 4 G 37790 in seed development. • Salik examination • MADISON FACILITY: Continue to search for the knockout by narrowing DNA Pool 4 down to a specific seed pool and then down to a specific t-DNA insert line. When this line is found, grow plants and continue to investigate whether knocking out AT 4 G 37790 significantly affects seed development.

How do we Study Gene Activity • In the SRB? 1. Isolate RNA from different vegetative organs of the SRB. 2. Remove contamination from RNA (such as genomic DNA). 3. Perform RT-PCR to transform m. RNA into c. DNA, then amplify c. DNA using primers for the genespecific region. 4. RT-PCR Methodology 5. Run products on a gel to see which samples amplified. The organs that provided samples with the most amplification are the ones where the gene is most active.