Knockout Mouse Project KOMP and Knockout Mouse Production




- Slides: 26

Knockout Mouse Project (KOMP) and Knockout Mouse Production and Phenotyping (KOMP 2) Mouse 101 Oct 19, 2015

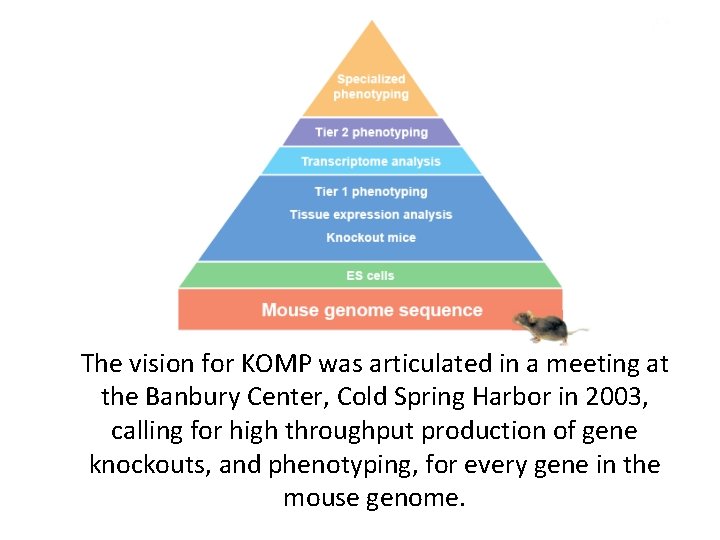
The vision for KOMP was articulated in a meeting at the Banbury Center, Cold Spring Harbor in 2003, calling for high throughput production of gene knockouts, and phenotyping, for every gene in the mouse genome.

Deltagen/Lexicon Lines http: //www. informatics. jax. org/external/ko/

KOMP (2006 -2011) • “…a high-throughput international effort to produce…knockouts for all mouse genes, and place these resources into the public domain. ” • The KOMP was launched in 2006 by NIH – $56. 6 million over 5 years from the ICs – a goal of creating 8, 500 ES cell lines – alleles are nulls or conditional-ready, contain reporter • The EC launched EUCOMM, the European Conditional Mouse Mutagenesis Program in October 2005 (funded in Feb 2005) – 13 M Euros over 3 years – a goal of creating 8, 000 mutants. • KOMP and EUCOMM along with other international efforts formed the International Knockout Mouse Consortium (IKMC) and have jointly produced > 17, 000 mutant ES cell lines and made them available from public repositories.

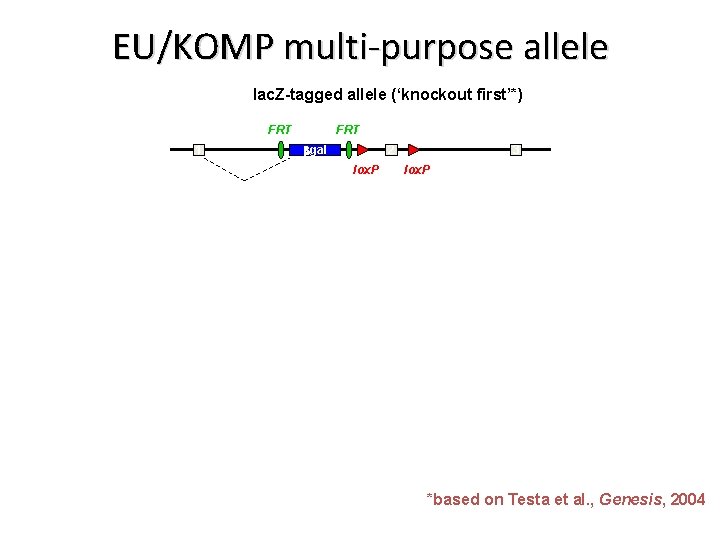
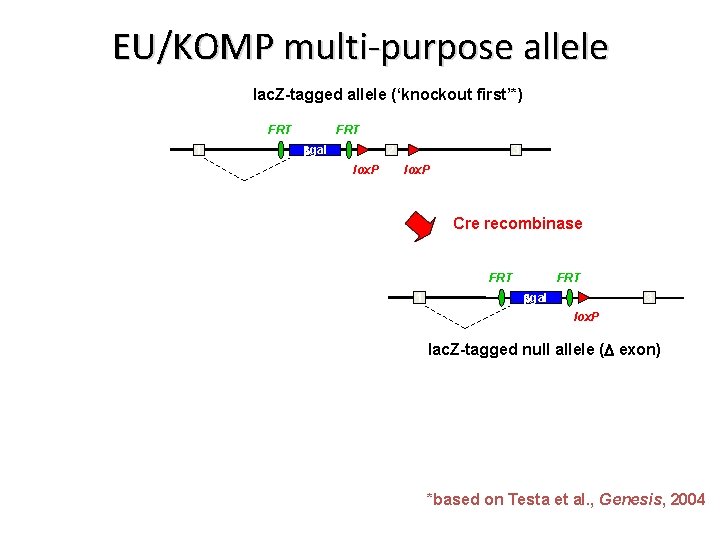
EU/KOMP multi-purpose allele lac. Z-tagged allele (‘knockout first’*) FRT 1 FRT bgal 2 lox. P 3 lox. P *based on Testa et al. , Genesis, 2004

EU/KOMP multi-purpose allele lac. Z-tagged allele (‘knockout first’*) FRT 1 FRT bgal 2 lox. P 3 lox. P Cre recombinase FRT 1 FRT bgal 3 lox. P lac. Z-tagged null allele (D exon) *based on Testa et al. , Genesis, 2004

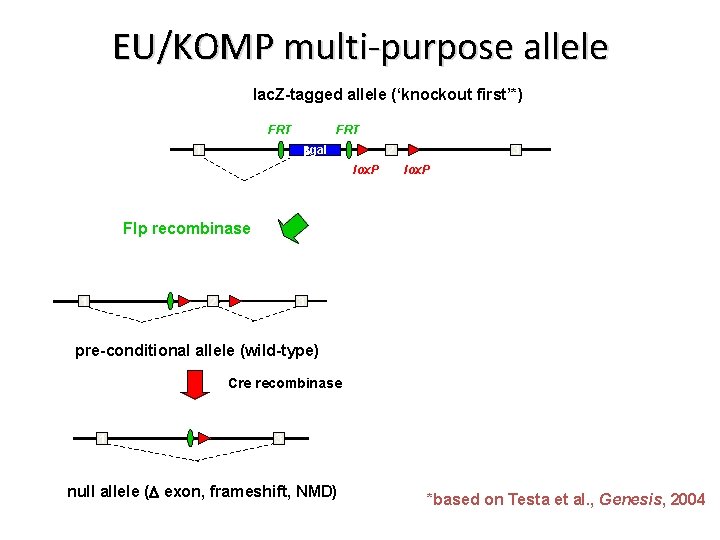
EU/KOMP multi-purpose allele lac. Z-tagged allele (‘knockout first’*) FRT bgal 1 2 lox. P 3 lox. P Flp recombinase 1 2 3 pre-conditional allele (wild-type) Cre recombinase 1 3 null allele (D exon, frameshift, NMD) *based on Testa et al. , Genesis, 2004

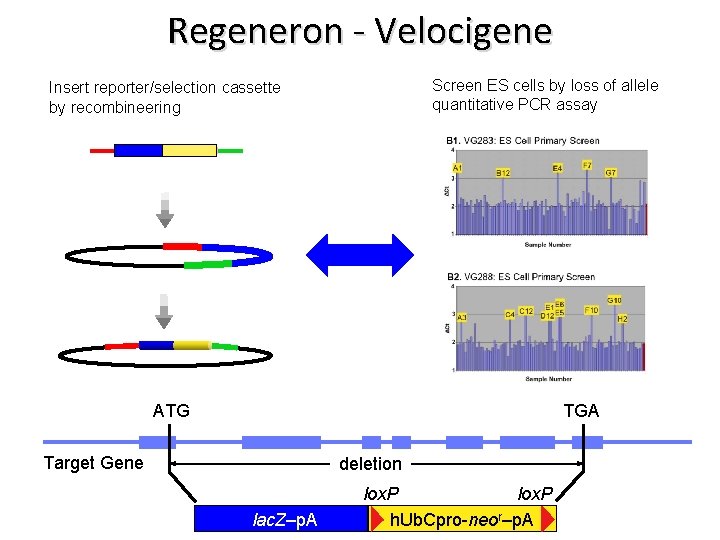
Regeneron - Velocigene Screen ES cells by loss of allele quantitative PCR assay Insert reporter/selection cassette by recombineering ATG TGA Target Gene deletion lac. Z–p. A lox. P h. Ub. Cpro-neor–p. A

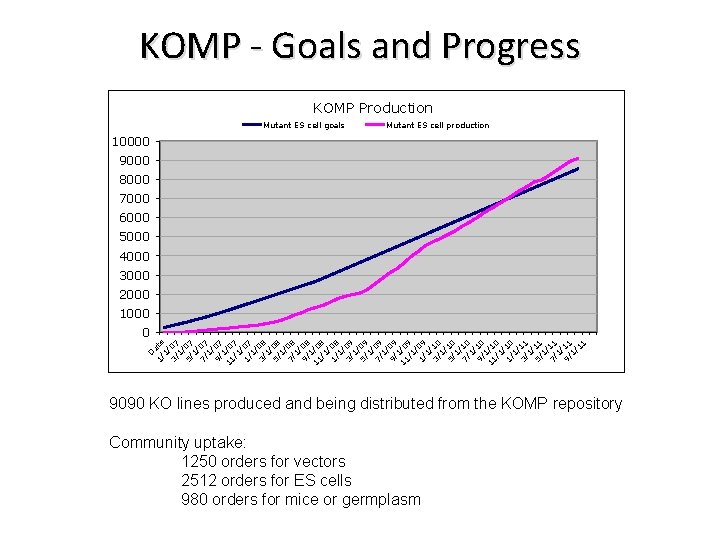
KOMP - Goals and Progress KOMP Production Mutant ES cell goals Mutant ES cell production 10000 9000 8000 7000 6000 5000 4000 3000 2000 1000 D 1/ ate 1/ 3/ 07 1/ 5/ 07 1/ 7/ 07 1/ 9/ 07 1 11 /0 /1 7 / 1/ 07 1/ 3/ 08 1/ 5/ 08 1/ 7/ 08 1/ 9/ 08 1 11 /0 /1 8 / 1/ 08 1/ 3/ 09 1/ 5/ 09 1/ 7/ 09 1/ 9/ 09 1 11 /0 /1 9 / 1/ 09 1/ 3/ 10 1/ 5/ 10 1/ 7/ 10 1/ 9/ 10 1 11 /1 /1 0 / 1/ 10 1/ 3/ 11 1/ 5/ 11 1/ 7/ 11 1/ 9/ 11 1/ 11 0 9090 KO lines produced and being distributed from the KOMP repository Community uptake: 1250 orders for vectors 2512 orders for ES cells 980 orders for mice or germplasm

KOMP 2 - Scientific Rationale • Provides access to unannotated genes by providing hypothesis testing and tools • Provides new insights into pleiotropy

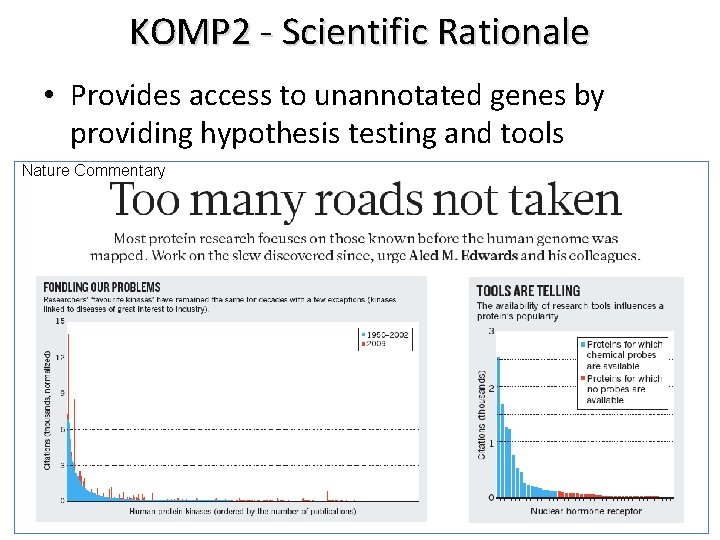
KOMP 2 - Scientific Rationale • Provides access to unannotated genes by providing hypothesis testing and tools Nature Commentary

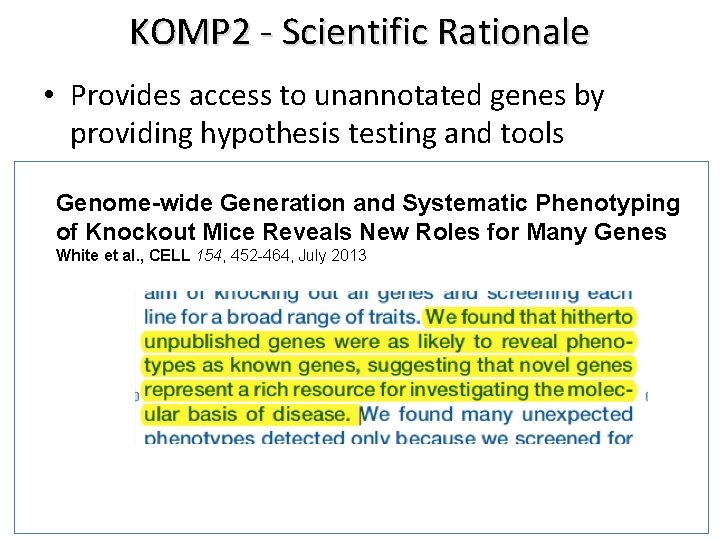
KOMP 2 - Scientific Rationale • Provides access to unannotated genes by providing hypothesis testing and tools Genome-wide Generation and Systematic Phenotyping of Knockout Mice Reveals New Roles for Many Genes White et al. , CELL 154, 452 -464, July 2013

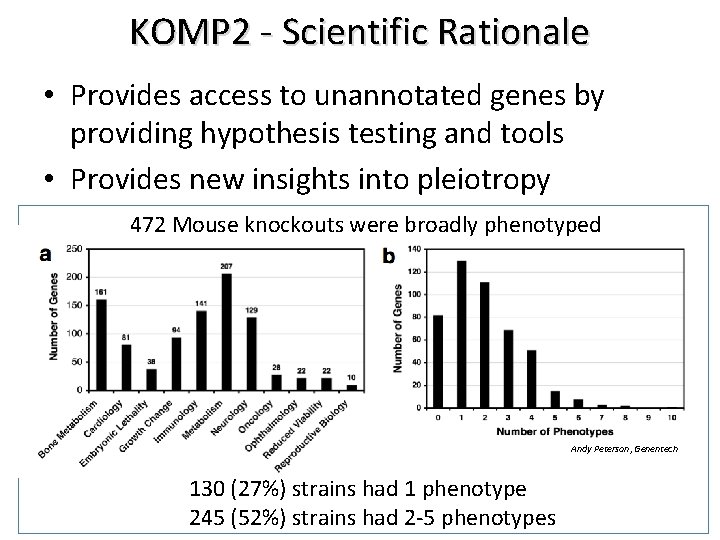
KOMP 2 - Scientific Rationale • Provides access to unannotated genes by providing hypothesis testing and tools • Provides new insights into pleiotropy 472 Mouse knockouts were broadly phenotyped Andy Peterson, Genentech 130 (27%) strains had 1 phenotype 245 (52%) strains had 2 -5 phenotypes

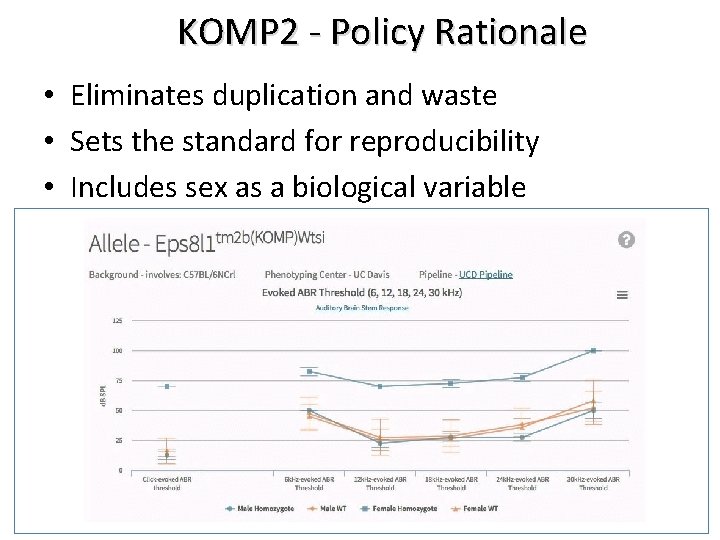
KOMP 2 - Policy Rationale • Eliminates duplication and waste • Sets the standard for reproducibility • Includes sex as a biological variable

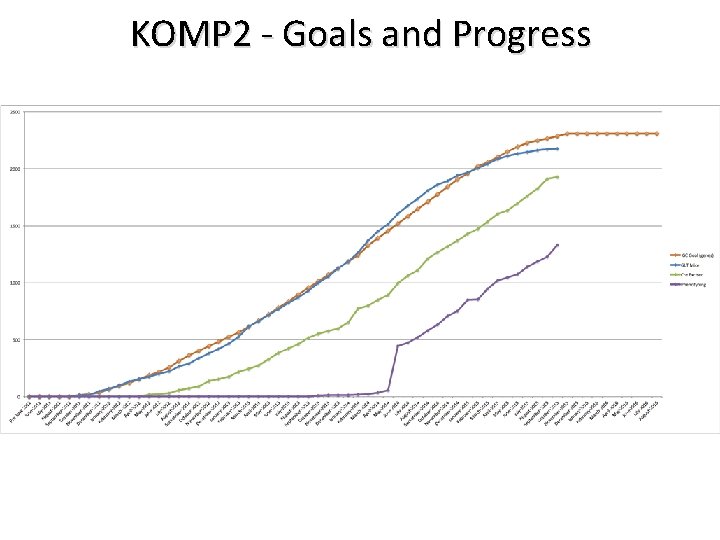
KOMP 2 Project Goals - (2011 -2021) • • Phase 1 (2011 -2016): Phenotype up to 2, 500 lines – Pipeline development, logistics – Phenotype technology developments – Economies of scale Phase 2 (2016 -2021): Phenotype 6, 000 mutants – Business plan in preparation • Data freely available through a Data Coordination Center – “One stop shop” Web Portal • Mice available through the global network of mouse repositories • Coordinate with IMPC to achieve broad-based phenotyping of 20, 000 mutants from the IKMC resource – A collaborative activity of mouse centers worldwide

KOMP 2 - Goals and Progress

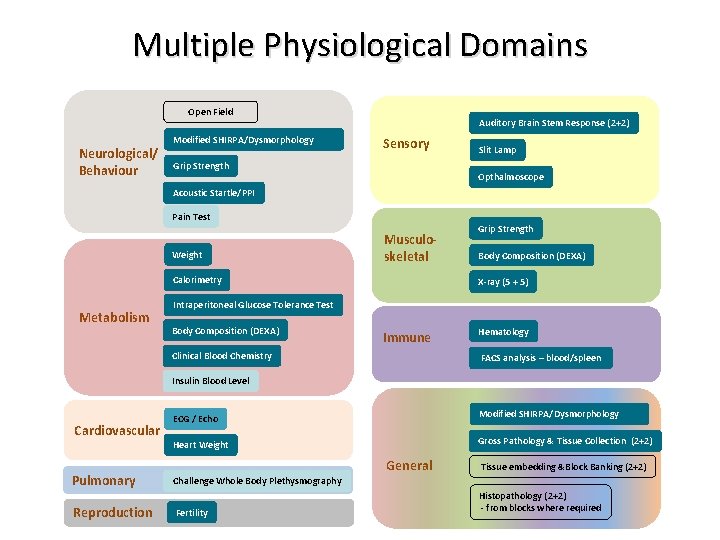
Multiple Physiological Domains Open Field Neurological/ Behaviour Modified SHIRPA/Dysmorphology Auditory Brain Stem Response (2+2) Sensory Grip Strength Slit Lamp Opthalmoscope Acoustic Startle/PPI Pain Test Weight Musculoskeletal Calorimetry Metabolism Grip Strength Body Composition (DEXA) X-ray (5 + 5) Intraperitoneal Glucose Tolerance Test Body Composition (DEXA) Immune Clinical Blood Chemistry Hematology FACS analysis – blood/spleen Insulin Blood Level Cardiovascular Pulmonary Reproduction ECG / Echo Modified SHIRPA/Dysmorphology Heart Weight Gross Pathology & Tissue Collection (2+2) General Tissue embedding &Block Banking (2+2) Challenge Whole Body Plethysmography Fertility Histopathology (2+2) - from blocks where required

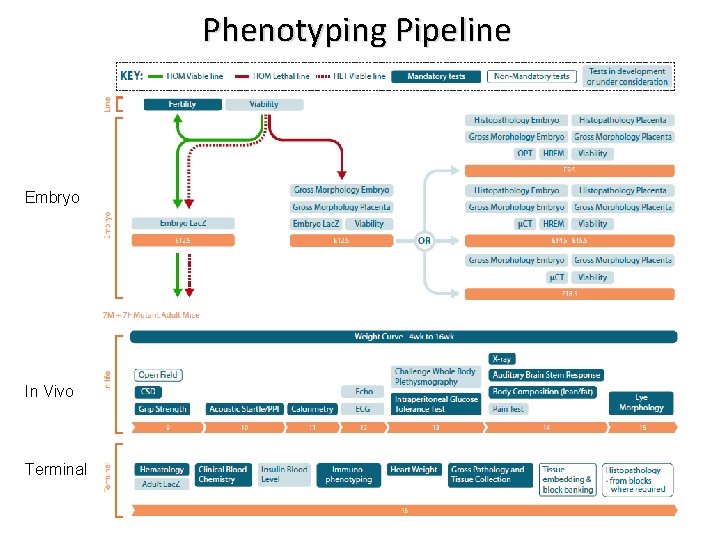
Phenotyping Pipeline Embryo In Vivo Terminal

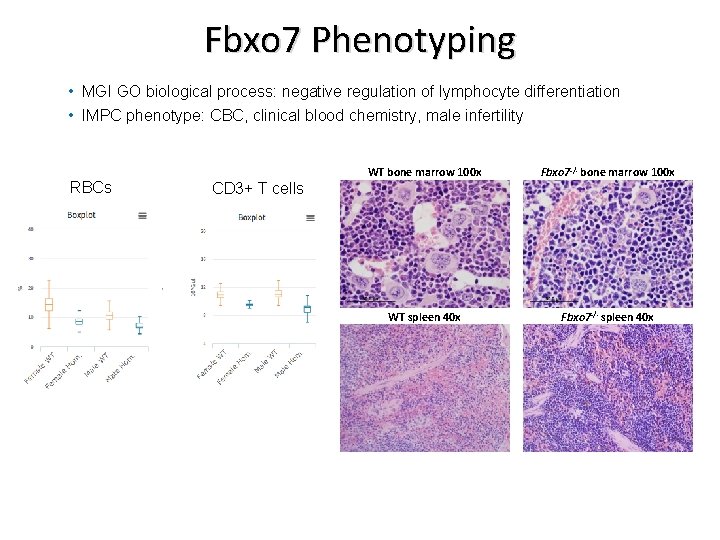
Fbxo 7 Phenotyping • MGI GO biological process: negative regulation of lymphocyte differentiation • IMPC phenotype: CBC, clinical blood chemistry, male infertility RBCs WT bone marrow 100 x Fbxo 7 -/- bone marrow 100 x WT spleen 40 x Fbxo 7 -/- spleen 40 x CD 3+ T cells

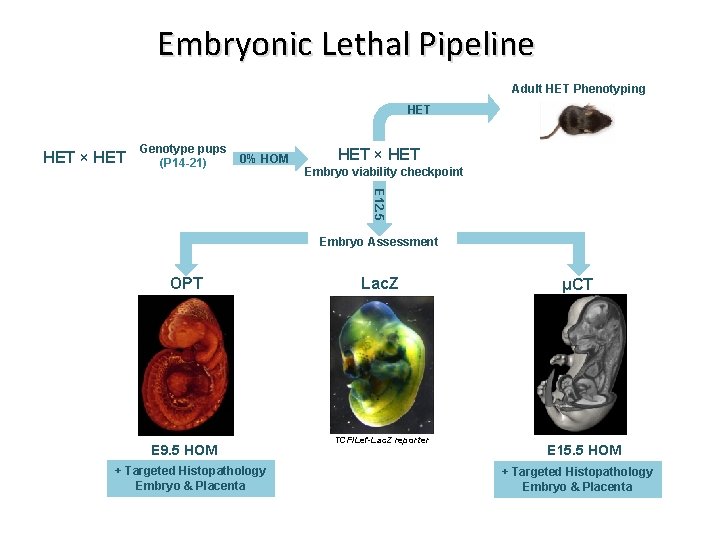
Embryonic Lethal Pipeline Adult HET Phenotyping HET × HET Genotype pups (P 14 -21) 0% HOM HET × HET Embryo viability checkpoint E 12. 5 Embryo Assessment OPT E 9. 5 HOM + Targeted Histopathology Embryo & Placenta Lac. Z TCF/Lef-Lac. Z reporter μCT E 15. 5 HOM + Targeted Histopathology Embryo & Placenta

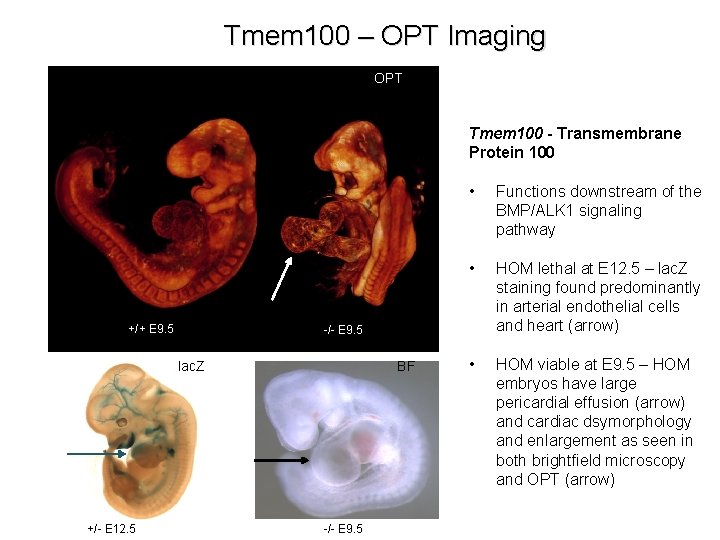
Tmem 100 – OPT Imaging OPT Tmem 100 - Transmembrane Protein 100 +/+ E 9. 5 Functions downstream of the BMP/ALK 1 signaling pathway • HOM lethal at E 12. 5 – lac. Z staining found predominantly in arterial endothelial cells and heart (arrow) • HOM viable at E 9. 5 – HOM embryos have large pericardial effusion (arrow) and cardiac dsymorphology and enlargement as seen in both brightfield microscopy and OPT (arrow) -/- E 9. 5 lac. Z +/- E 12. 5 • BF -/- E 9. 5

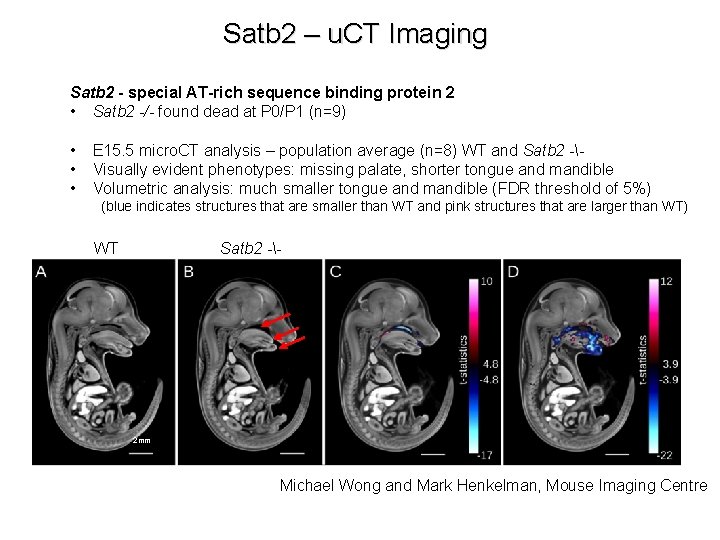
Satb 2 – u. CT Imaging Satb 2 - special AT-rich sequence binding protein 2 • Satb 2 -/- found dead at P 0/P 1 (n=9) • • • E 15. 5 micro. CT analysis – population average (n=8) WT and Satb 2 -Visually evident phenotypes: missing palate, shorter tongue and mandible Volumetric analysis: much smaller tongue and mandible (FDR threshold of 5%) (blue indicates structures that are smaller than WT and pink structures that are larger than WT) WT Satb 2 -- 2 mm Michael Wong and Mark Henkelman, Mouse Imaging Centre

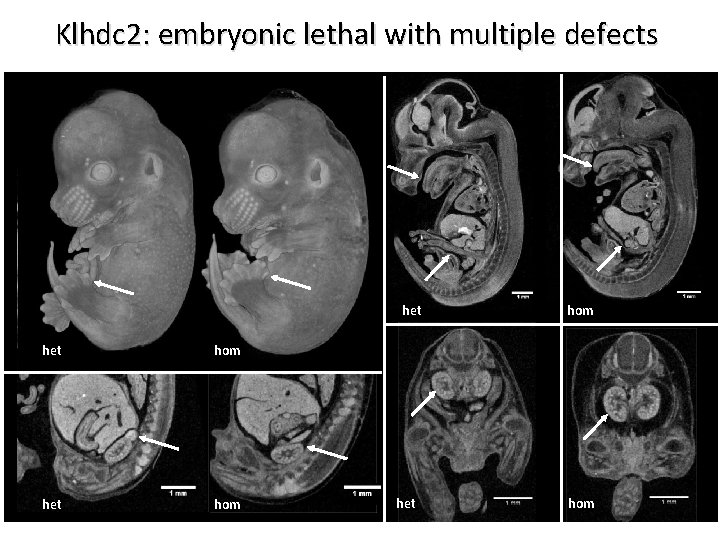
Klhdc 2: embryonic lethal with multiple defects het hom

Database Access

www. mousephenotype. org

Korean Mouse Phenotyping Centre